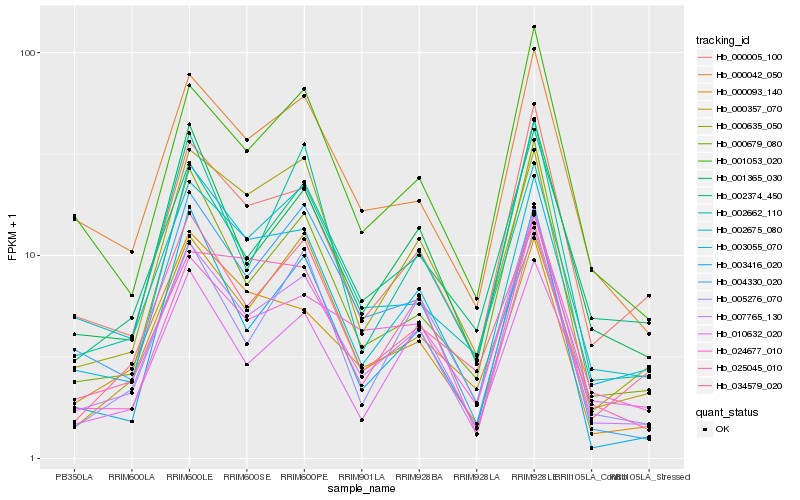
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_130 |
0.0 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000042_050 |
0.0937400463 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003055_070 |
0.0953415202 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001365_030 |
0.0966303958 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 5 |
Hb_000005_100 |
0.1068500528 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000679_080 |
0.1089847113 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000093_140 |
0.11006291 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000635_050 |
0.1114489789 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 9 |
Hb_002374_450 |
0.1123019119 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001053_020 |
0.1126141914 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 11 |
Hb_002675_080 |
0.1148650011 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 12 |
Hb_003416_020 |
0.1152503423 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 13 |
Hb_005276_070 |
0.1157820505 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 14 |
Hb_025045_010 |
0.1174720522 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_002662_110 |
0.1182873885 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_000357_070 |
0.1210143025 |
- |
- |
- |
| 17 |
Hb_024677_010 |
0.1213359833 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 18 |
Hb_010632_020 |
0.1222442901 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 19 |
Hb_034579_020 |
0.1230219598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004330_020 |
0.1264832263 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |