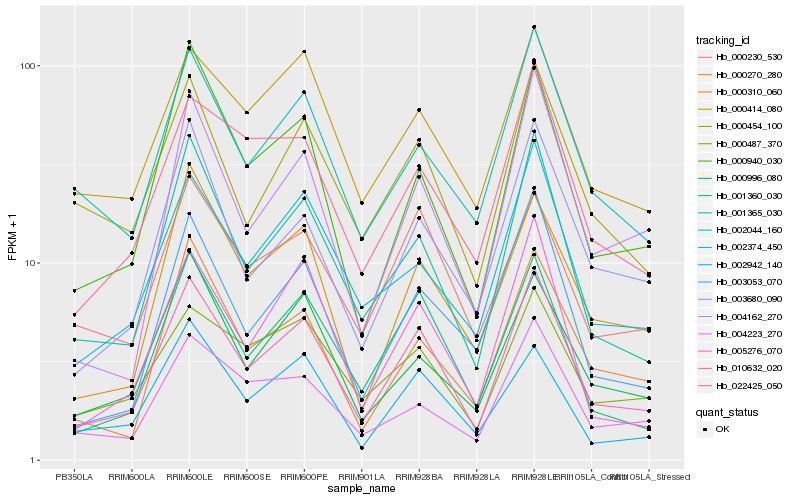
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010632_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 2 |
Hb_001365_030 |
0.0854949057 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 3 |
Hb_001360_030 |
0.0923295264 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 4 |
Hb_000940_030 |
0.1076851013 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002374_450 |
0.1101811586 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003053_070 |
0.110797387 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005276_070 |
0.1129046284 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 8 |
Hb_002044_160 |
0.1138946774 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 9 |
Hb_000310_060 |
0.114612851 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 10 |
Hb_000454_100 |
0.1151809966 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 11 |
Hb_000996_080 |
0.1163488148 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 12 |
Hb_003680_090 |
0.116458652 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 13 |
Hb_022425_050 |
0.1177371038 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 14 |
Hb_004162_270 |
0.1181732512 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_002942_140 |
0.1194469516 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 16 |
Hb_000270_280 |
0.1209425763 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_000414_080 |
0.1215060274 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000230_530 |
0.1216912918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004223_270 |
0.121960342 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000487_370 |
0.1221656905 |
- |
- |
- |