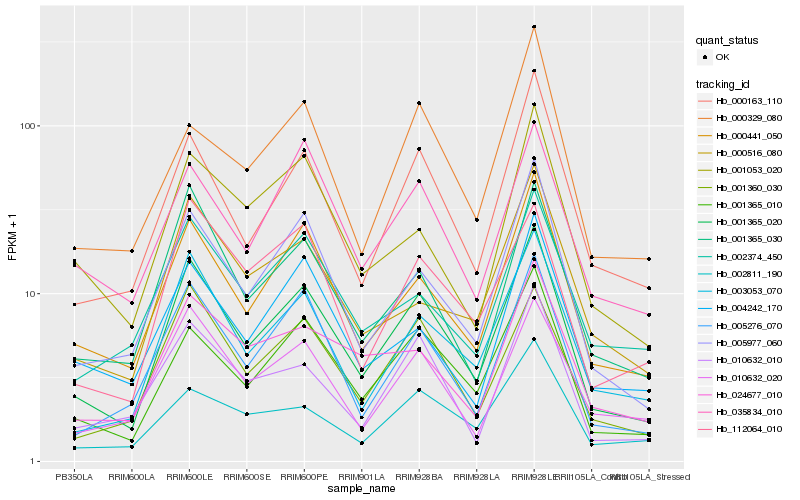
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001365_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 2 |
Hb_001365_010 |
0.0875499912 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 3 |
Hb_000441_050 |
0.0938603411 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 4 |
Hb_003053_070 |
0.0992777563 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005977_060 |
0.1003060616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000163_110 |
0.1006418092 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 7 |
Hb_005276_070 |
0.1053856469 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 8 |
Hb_001360_030 |
0.1108914036 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 9 |
Hb_001365_030 |
0.1141635199 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 10 |
Hb_010632_010 |
0.1230433247 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_010632_020 |
0.1235098707 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 12 |
Hb_024677_010 |
0.1322766143 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 13 |
Hb_035834_010 |
0.1334346078 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_112064_010 |
0.1334695969 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 15 |
Hb_000516_080 |
0.1335240928 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 16 |
Hb_001053_020 |
0.1345465472 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 17 |
Hb_002811_190 |
0.1357533014 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 18 |
Hb_002374_450 |
0.1374900422 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004242_170 |
0.1381199807 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 20 |
Hb_000329_080 |
0.1398866077 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |