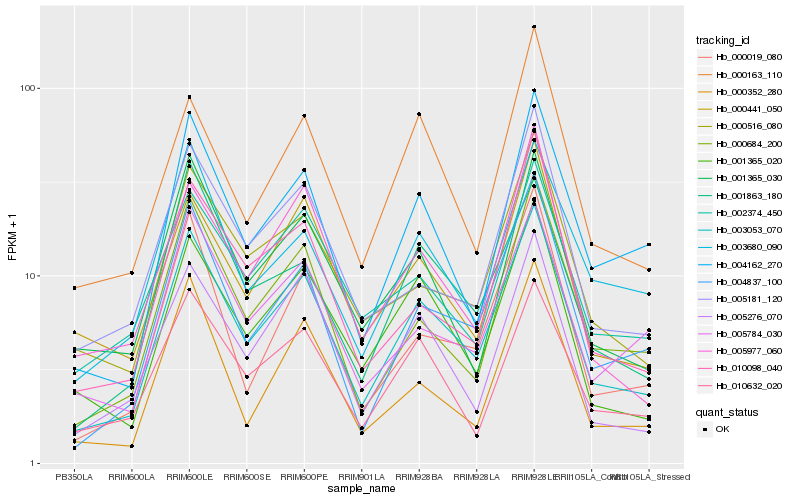
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003053_070 |
0.0 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004837_100 |
0.0933955907 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001365_020 |
0.0992777563 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 4 |
Hb_000684_200 |
0.1002867901 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_004162_270 |
0.1025571619 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000516_080 |
0.1088399687 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 7 |
Hb_001365_030 |
0.1092650894 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 8 |
Hb_010632_020 |
0.110797387 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 9 |
Hb_005276_070 |
0.1109715025 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 10 |
Hb_001863_180 |
0.1128487784 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000019_080 |
0.1134426313 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 12 |
Hb_005784_030 |
0.1148435532 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000441_050 |
0.1158355975 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 14 |
Hb_003680_090 |
0.1162616349 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005977_060 |
0.1176972243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000163_110 |
0.1181877222 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 17 |
Hb_005181_120 |
0.1192601511 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_010098_040 |
0.1194116375 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 19 |
Hb_002374_450 |
0.1216580379 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000352_280 |
0.1218770412 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |