| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
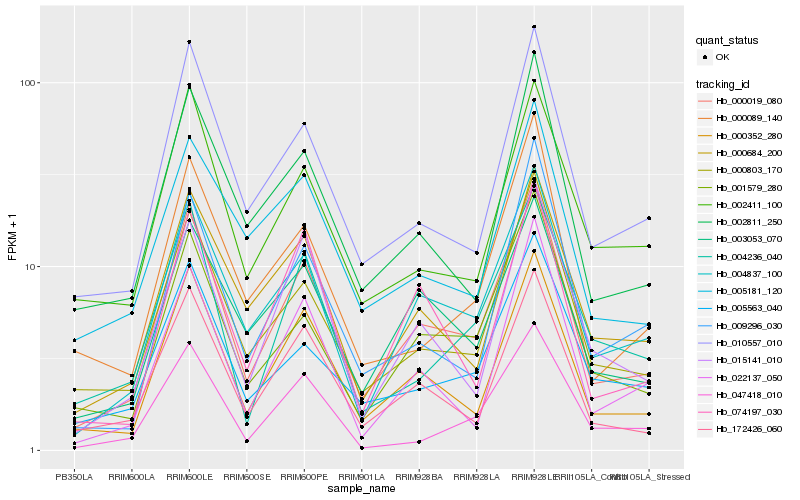
Hb_000019_080 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 2 |
Hb_004837_100 |
0.0790713474 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000352_280 |
0.0795149664 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 4 |
Hb_000803_170 |
0.0945171412 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 5 |
Hb_172426_060 |
0.0945825941 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 6 |
Hb_001579_280 |
0.1013543163 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA synthase, peroxisomal-like [Jatropha curcas] |
| 7 |
Hb_010557_010 |
0.1021409446 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002811_250 |
0.1106605368 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000684_200 |
0.1115019084 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_003053_070 |
0.1134426313 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005563_040 |
0.119662868 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 12 |
Hb_015141_010 |
0.1230203797 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 13 |
Hb_022137_050 |
0.1255388197 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 14 |
Hb_000089_140 |
0.1263187528 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004236_040 |
0.126577385 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 16 |
Hb_074197_030 |
0.1277555462 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002411_100 |
0.1289790727 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 18 |
Hb_009296_030 |
0.1294411427 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 19 |
Hb_047418_010 |
0.1303325894 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 20 |
Hb_005181_120 |
0.1316797675 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |