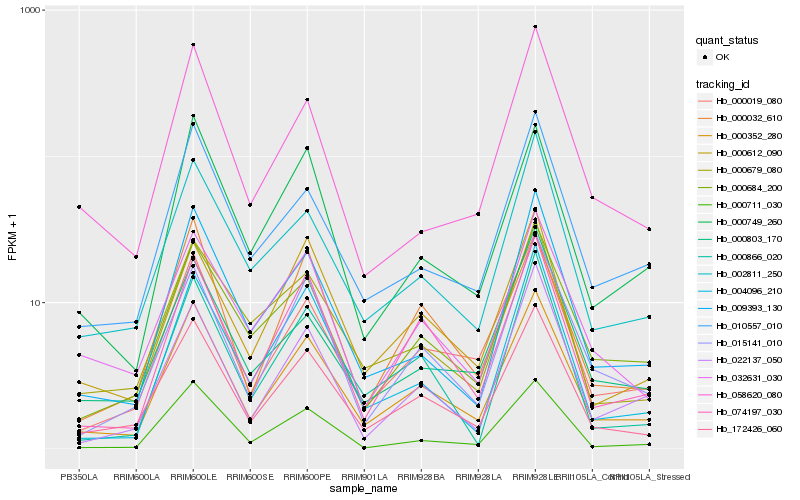
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_280 |
0.0 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 2 |
Hb_172426_060 |
0.0561805365 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 3 |
Hb_000019_080 |
0.0795149664 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 4 |
Hb_074197_030 |
0.0898180173 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_009393_130 |
0.0991540476 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 6 |
Hb_015141_010 |
0.0994785618 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 7 |
Hb_010557_010 |
0.099714074 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000684_200 |
0.101056246 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_002811_250 |
0.1019071327 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000749_260 |
0.1089793485 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000612_090 |
0.1128262875 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 12 |
Hb_000711_030 |
0.1171240352 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 13 |
Hb_032631_030 |
0.1183549952 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 14 |
Hb_000032_610 |
0.1188145573 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_022137_050 |
0.1192648678 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 16 |
Hb_004096_210 |
0.1192883564 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000803_170 |
0.1197161628 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 18 |
Hb_000679_080 |
0.1208888239 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000866_020 |
0.1210065574 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 20 |
Hb_058620_080 |
0.1213647537 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |