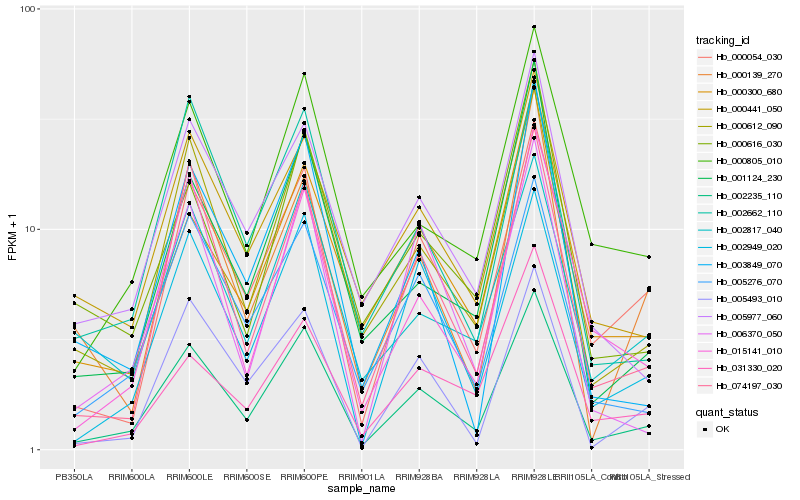
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_110 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 2 |
Hb_000612_090 |
0.0971397583 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 3 |
Hb_002949_020 |
0.1046832721 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 4 |
Hb_074197_030 |
0.106099381 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000054_030 |
0.108107783 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 6 |
Hb_005276_070 |
0.1158035113 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 7 |
Hb_000805_010 |
0.1199306637 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 8 |
Hb_015141_010 |
0.1218444816 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 9 |
Hb_005977_060 |
0.1258929258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002662_110 |
0.1305715898 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_001124_230 |
0.1312824043 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006370_050 |
0.1314540681 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_005493_010 |
0.1318595591 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_003849_070 |
0.1321357614 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 15 |
Hb_000441_050 |
0.1322134015 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 16 |
Hb_002817_040 |
0.1371744988 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 17 |
Hb_000616_030 |
0.1430302258 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 18 |
Hb_031330_020 |
0.1447058442 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 19 |
Hb_000139_270 |
0.1448159097 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 20 |
Hb_000300_680 |
0.1458195976 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |