| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
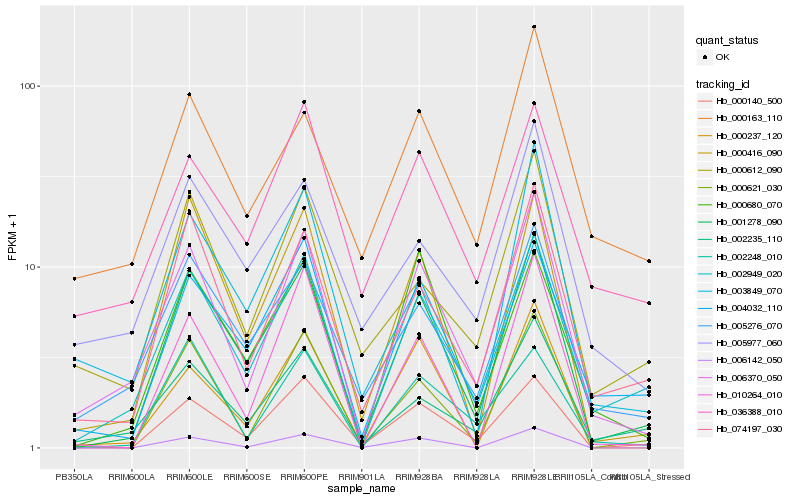
Hb_006370_050 |
0.0 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002949_020 |
0.1198508862 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 3 |
Hb_002235_110 |
0.1314540681 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 4 |
Hb_001278_090 |
0.1332765978 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 5 |
Hb_074197_030 |
0.1426236512 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_005276_070 |
0.1433969213 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 7 |
Hb_000621_030 |
0.1603142903 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000416_090 |
0.1627893026 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 9 |
Hb_000237_120 |
0.1667439045 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 10 |
Hb_000612_090 |
0.1667458693 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 11 |
Hb_005977_060 |
0.1673513503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000140_500 |
0.1682227912 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000163_110 |
0.1683437826 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 14 |
Hb_003849_070 |
0.1697642407 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 15 |
Hb_004032_110 |
0.1717338566 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002248_010 |
0.17474318 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 17 |
Hb_000680_070 |
0.1747980317 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006142_050 |
0.1751842737 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 19 |
Hb_036388_010 |
0.1753613987 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 20 |
Hb_010264_010 |
0.1756467296 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |