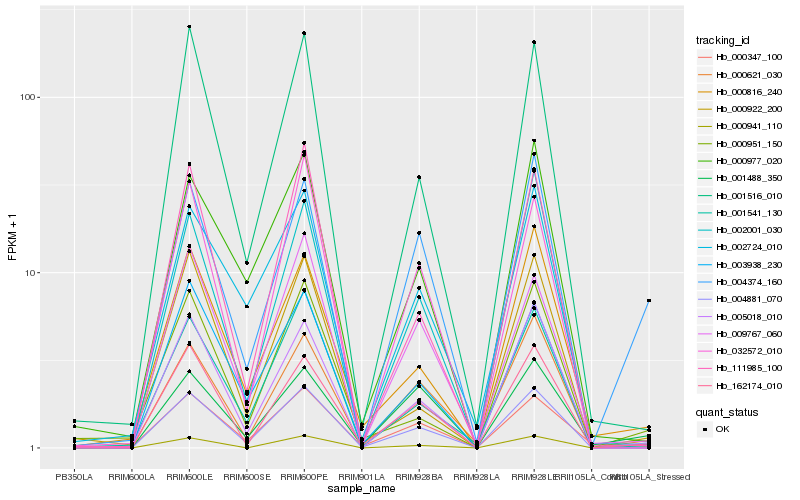
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_070 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_162174_010 |
0.1032073776 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000621_030 |
0.1057779554 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004374_160 |
0.1203644391 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 5 |
Hb_001541_130 |
0.1237499679 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 6 |
Hb_001516_010 |
0.1261360424 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 7 |
Hb_000816_240 |
0.126658997 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000347_100 |
0.1270206229 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 9 |
Hb_005018_010 |
0.1285866306 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_009767_060 |
0.1327106631 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 11 |
Hb_000951_150 |
0.1355686331 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 12 |
Hb_002001_030 |
0.1360935324 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 13 |
Hb_000977_020 |
0.1378372562 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 14 |
Hb_003938_230 |
0.1388933329 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 15 |
Hb_032572_010 |
0.1415355127 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 16 |
Hb_001488_350 |
0.1418185994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_111985_100 |
0.1419814097 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_000941_110 |
0.1425677081 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_002724_010 |
0.1428277402 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 20 |
Hb_000922_200 |
0.1436461873 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |