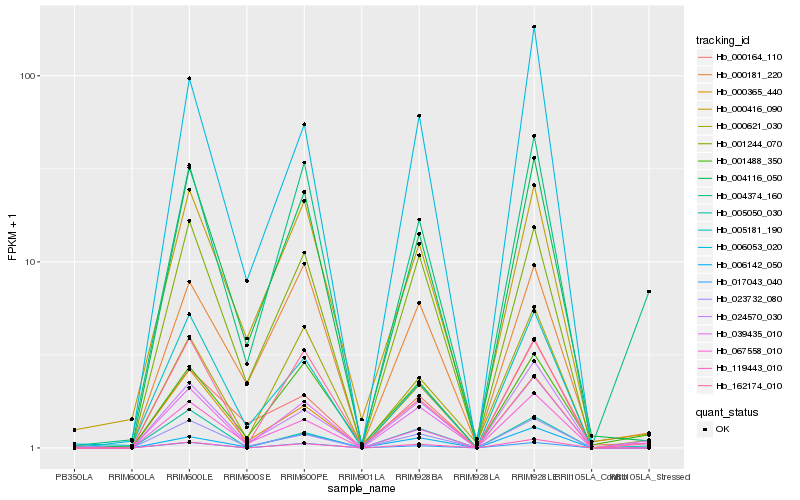
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039435_010 |
0.0 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 2 |
Hb_024570_030 |
0.0886935687 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_004374_160 |
0.1102959806 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 4 |
Hb_001488_350 |
0.1106801677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004116_050 |
0.1137457509 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 6 |
Hb_119443_010 |
0.1169855523 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 7 |
Hb_001244_070 |
0.1241897339 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 8 |
Hb_000621_030 |
0.1245636672 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 9 |
Hb_067558_010 |
0.1323614968 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_162174_010 |
0.1344919214 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_006053_020 |
0.1374138563 |
- |
- |
CAP [Theobroma cacao] |
| 12 |
Hb_005181_190 |
0.1408653939 |
- |
- |
PREDICTED: uncharacterized protein LOC105643550 [Jatropha curcas] |
| 13 |
Hb_006142_050 |
0.1416526419 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 14 |
Hb_000365_440 |
0.1437402294 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_005050_030 |
0.1443427926 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 16 |
Hb_017043_040 |
0.1445014268 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000416_090 |
0.1486853943 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 18 |
Hb_023732_080 |
0.1489759189 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 19 |
Hb_000181_220 |
0.1498036745 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000164_110 |
0.1504433376 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |