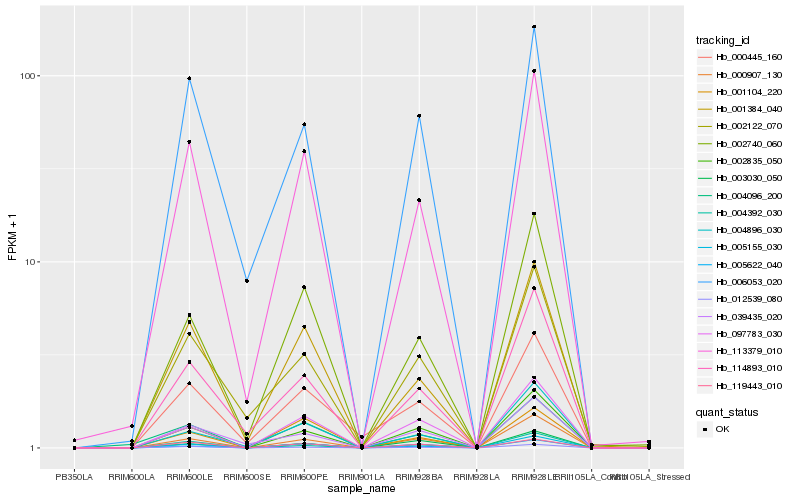
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005155_030 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_097783_030 |
0.0534728349 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 3 |
Hb_002835_050 |
0.0544382888 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_000907_130 |
0.0662546837 |
- |
- |
- |
| 5 |
Hb_003030_050 |
0.0915494613 |
- |
- |
- |
| 6 |
Hb_113379_010 |
0.1008352074 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 7 |
Hb_002740_060 |
0.1015909579 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 8 |
Hb_012539_080 |
0.1084500156 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 9 |
Hb_005622_040 |
0.1158995814 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 10 |
Hb_119443_010 |
0.1228750477 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 11 |
Hb_114893_010 |
0.1257855892 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 12 |
Hb_001384_040 |
0.1262416436 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 13 |
Hb_004896_030 |
0.1287695895 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 14 |
Hb_006053_020 |
0.1335309251 |
- |
- |
CAP [Theobroma cacao] |
| 15 |
Hb_004096_200 |
0.1356875744 |
- |
- |
PREDICTED: GDT1-like protein 5 [Eucalyptus grandis] |
| 16 |
Hb_000445_160 |
0.1359799498 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 17 |
Hb_002122_070 |
0.1378247955 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_039435_020 |
0.1384649713 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_004392_030 |
0.1385636668 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 20 |
Hb_001104_220 |
0.1441368827 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |