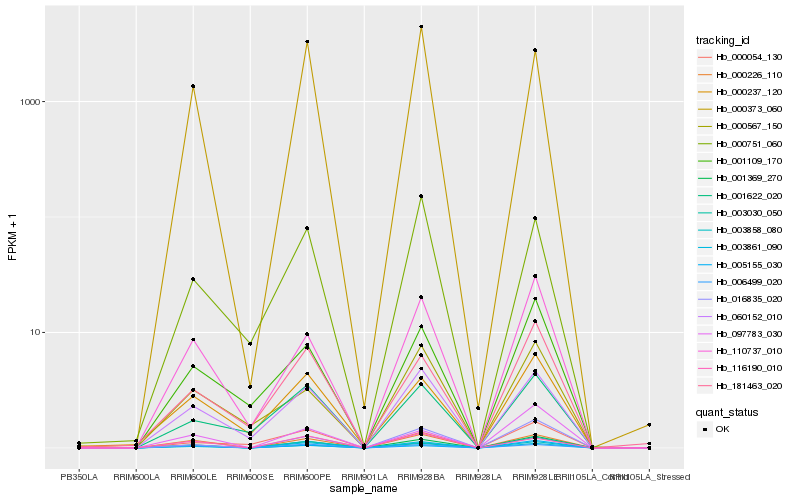
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_270 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000226_110 |
0.1080839818 |
- |
- |
- |
| 3 |
Hb_110737_010 |
0.1137953137 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 4 |
Hb_016835_020 |
0.114713969 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_060152_010 |
0.1199597255 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 6 |
Hb_003858_080 |
0.122495841 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 7 |
Hb_003030_050 |
0.1372143397 |
- |
- |
- |
| 8 |
Hb_003861_090 |
0.1411914428 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 9 |
Hb_001109_170 |
0.1435554333 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_181463_020 |
0.1438352776 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 11 |
Hb_097783_030 |
0.1464260664 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 12 |
Hb_000237_120 |
0.1488246753 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 13 |
Hb_006499_020 |
0.1489423497 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_001622_020 |
0.1507168307 |
- |
- |
- |
| 15 |
Hb_000373_060 |
0.154493114 |
- |
- |
PREDICTED: extensin-3 [Camelina sativa] |
| 16 |
Hb_000054_130 |
0.1676335643 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 17 |
Hb_005155_030 |
0.1678804379 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 18 |
Hb_000751_060 |
0.1695343043 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 19 |
Hb_116190_010 |
0.1699936287 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 20 |
Hb_000567_150 |
0.170494417 |
- |
- |
cysteine protease, putative [Ricinus communis] |