| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
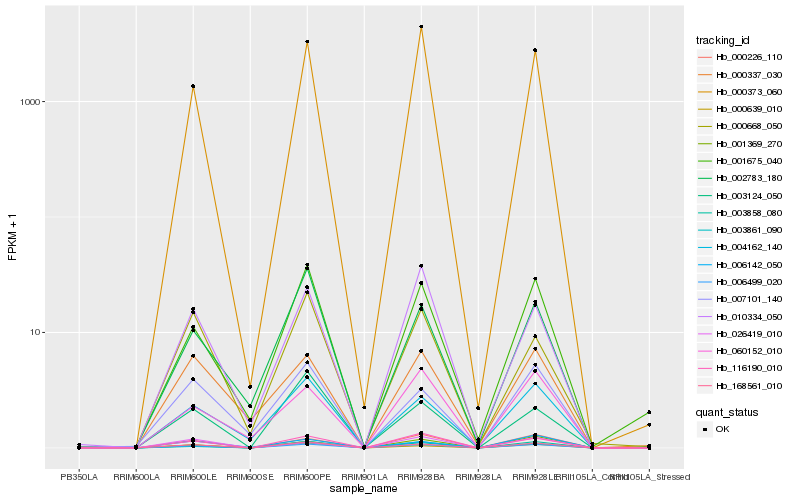
Hb_006499_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_168561_010 |
0.0502611945 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 3 |
Hb_116190_010 |
0.083968087 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 4 |
Hb_000226_110 |
0.0932246429 |
- |
- |
- |
| 5 |
Hb_003861_090 |
0.0951026977 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 6 |
Hb_000373_060 |
0.1100068668 |
- |
- |
PREDICTED: extensin-3 [Camelina sativa] |
| 7 |
Hb_000639_010 |
0.1184922748 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 8 |
Hb_003858_080 |
0.1271436801 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 9 |
Hb_004162_140 |
0.1315616195 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 10 |
Hb_001675_040 |
0.1357531445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001369_270 |
0.1489423497 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_010334_050 |
0.1495891896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_060152_010 |
0.1518249253 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 14 |
Hb_007101_140 |
0.154829751 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 15 |
Hb_000337_030 |
0.155801347 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_002783_180 |
0.1595250996 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 17 |
Hb_000668_050 |
0.161551855 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 18 |
Hb_006142_050 |
0.1636091036 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 19 |
Hb_026419_010 |
0.1649133276 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 20 |
Hb_003124_050 |
0.167224761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |