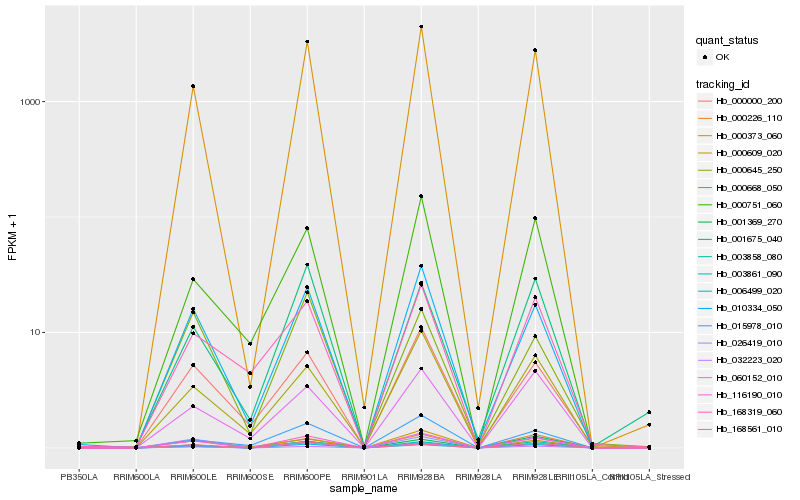
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116190_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 2 |
Hb_000373_060 |
0.0497919889 |
- |
- |
PREDICTED: extensin-3 [Camelina sativa] |
| 3 |
Hb_003861_090 |
0.0578603454 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 4 |
Hb_006499_020 |
0.083968087 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_010334_050 |
0.0850659392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000226_110 |
0.0993442294 |
- |
- |
- |
| 7 |
Hb_168561_010 |
0.1277233721 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 8 |
Hb_000000_200 |
0.1324781818 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 9 |
Hb_000645_250 |
0.1428718062 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 10 |
Hb_060152_010 |
0.1435434242 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 11 |
Hb_000609_020 |
0.147391585 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 12 |
Hb_003858_080 |
0.1528250326 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 13 |
Hb_000751_060 |
0.1580640302 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 14 |
Hb_032223_020 |
0.1598364322 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_001675_040 |
0.1623727374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_026419_010 |
0.163027405 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 17 |
Hb_000668_050 |
0.1664057548 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 18 |
Hb_168319_060 |
0.1667250743 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 19 |
Hb_015978_010 |
0.1680893698 |
- |
- |
PREDICTED: probable beta-D-xylosidase 2 [Populus euphratica] |
| 20 |
Hb_001369_270 |
0.1699936287 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |