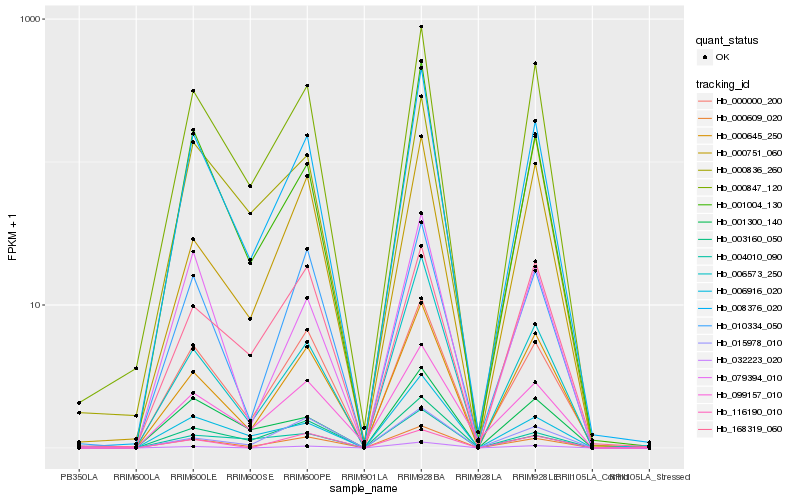
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000609_020 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 2 |
Hb_000000_200 |
0.0544508639 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 3 |
Hb_008376_020 |
0.0587038555 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 4 |
Hb_000847_120 |
0.0797885525 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 5 |
Hb_010334_050 |
0.0901501255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000645_250 |
0.0991612504 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 7 |
Hb_099157_010 |
0.1109807622 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 8 |
Hb_000836_260 |
0.1111909892 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_003160_050 |
0.1244105205 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_001004_130 |
0.1279754825 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 11 |
Hb_006573_250 |
0.1336358425 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 12 |
Hb_168319_060 |
0.1343902093 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 13 |
Hb_032223_020 |
0.1360112216 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001300_140 |
0.1363650862 |
- |
- |
- |
| 15 |
Hb_000751_060 |
0.1370550594 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 16 |
Hb_004010_090 |
0.137176643 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_116190_010 |
0.147391585 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 18 |
Hb_015978_010 |
0.1477179685 |
- |
- |
PREDICTED: probable beta-D-xylosidase 2 [Populus euphratica] |
| 19 |
Hb_079394_010 |
0.1478367895 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_006916_020 |
0.1483939823 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |