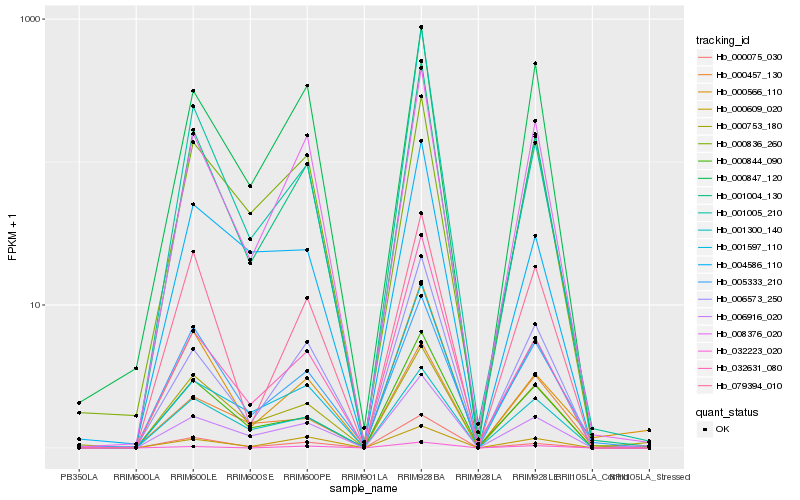
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001004_130 |
0.0 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 2 |
Hb_000844_090 |
0.0801570984 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 3 |
Hb_008376_020 |
0.0862300299 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 4 |
Hb_006573_250 |
0.091450106 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 5 |
Hb_079394_010 |
0.1023691457 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_001005_210 |
0.1054245583 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 7 |
Hb_001300_140 |
0.1118240191 |
- |
- |
- |
| 8 |
Hb_006916_020 |
0.1128589712 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 9 |
Hb_005333_210 |
0.114481346 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 10 |
Hb_032631_080 |
0.1174085709 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000609_020 |
0.1279754825 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 12 |
Hb_001597_110 |
0.1307584646 |
- |
- |
- |
| 13 |
Hb_000847_120 |
0.1315104394 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 14 |
Hb_000075_030 |
0.1376872043 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 15 |
Hb_032223_020 |
0.1398124758 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000753_180 |
0.1400607226 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 17 |
Hb_000457_130 |
0.1405735272 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |
| 18 |
Hb_000566_110 |
0.1417155768 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 19 |
Hb_004586_110 |
0.1431764515 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000836_260 |
0.1470378679 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |