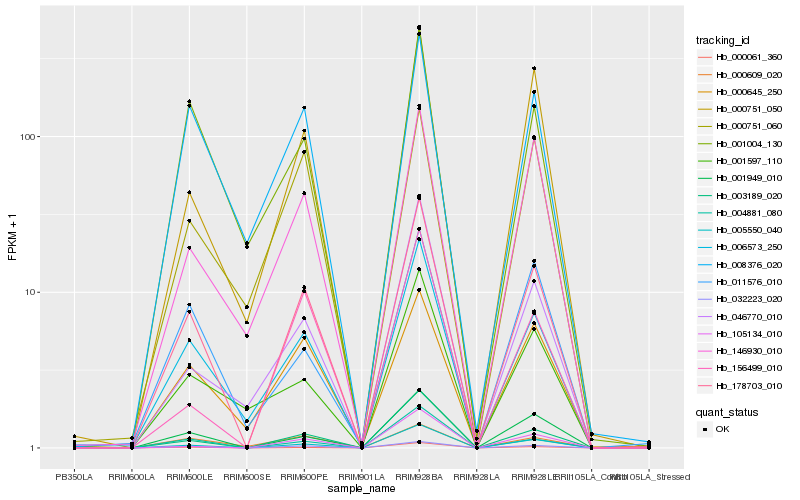
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178703_010 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_032223_020 |
0.0475596739 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_005550_040 |
0.0866859256 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 4 |
Hb_006573_250 |
0.0901600339 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 5 |
Hb_003189_020 |
0.0943323015 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 6 |
Hb_001949_010 |
0.1039859908 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 7 |
Hb_000751_050 |
0.1305871532 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 8 |
Hb_000061_360 |
0.1358744968 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 9 |
Hb_004881_080 |
0.1394150963 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 10 |
Hb_000645_250 |
0.1422639616 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 11 |
Hb_146930_010 |
0.1432215292 |
- |
- |
- |
| 12 |
Hb_008376_020 |
0.1438112562 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 13 |
Hb_105134_010 |
0.1485259829 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 14 |
Hb_046770_010 |
0.1492441309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001004_130 |
0.1522056603 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 16 |
Hb_001597_110 |
0.1551320624 |
- |
- |
- |
| 17 |
Hb_156499_010 |
0.1614558168 |
- |
- |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 18 |
Hb_000609_020 |
0.1673499352 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 19 |
Hb_011576_010 |
0.1682394984 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_000751_060 |
0.169532168 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |