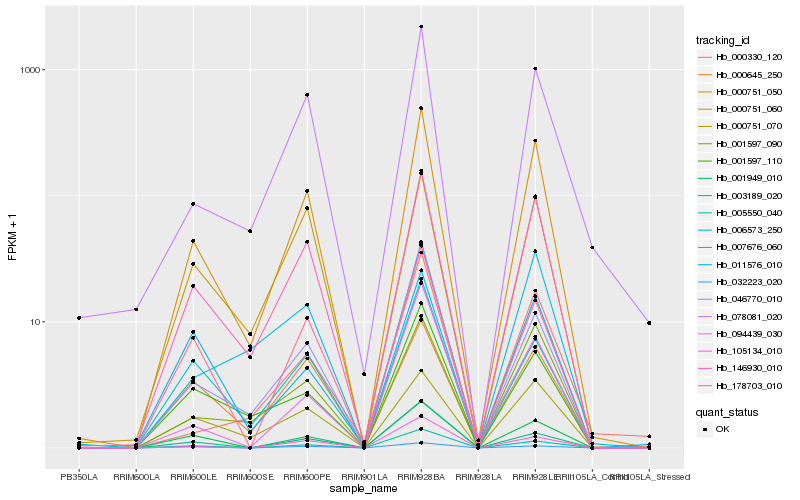
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000751_050 |
0.0 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 2 |
Hb_146930_010 |
0.0539905375 |
- |
- |
- |
| 3 |
Hb_001597_090 |
0.1142451748 |
- |
- |
- |
| 4 |
Hb_005550_040 |
0.1198866825 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 5 |
Hb_078081_020 |
0.1242047238 |
- |
- |
- |
| 6 |
Hb_001949_010 |
0.1243288625 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 7 |
Hb_178703_010 |
0.1305871532 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_046770_010 |
0.136102594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000751_070 |
0.1395887397 |
- |
- |
- |
| 10 |
Hb_105134_010 |
0.1417601728 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 11 |
Hb_001597_110 |
0.1455246509 |
- |
- |
- |
| 12 |
Hb_006573_250 |
0.1504660958 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 13 |
Hb_000751_060 |
0.1518922003 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 14 |
Hb_000330_120 |
0.1527046036 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 15 |
Hb_007676_060 |
0.1551960463 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 16 |
Hb_094439_030 |
0.1561854009 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 17 |
Hb_000645_250 |
0.1563359506 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 18 |
Hb_003189_020 |
0.1570539773 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 19 |
Hb_032223_020 |
0.1578782133 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_011576_010 |
0.1647329341 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |