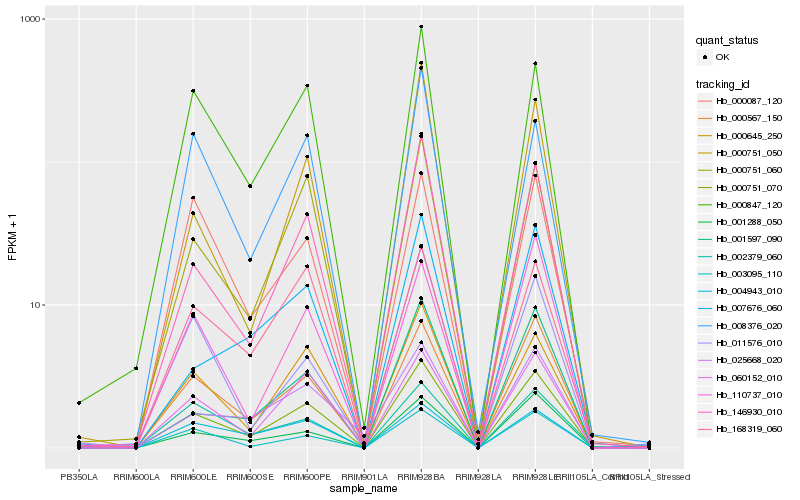
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000751_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000567_150 |
0.0753871775 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 3 |
Hb_146930_010 |
0.0940737974 |
- |
- |
- |
| 4 |
Hb_060152_010 |
0.0949161341 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_000645_250 |
0.0984378534 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 6 |
Hb_000751_060 |
0.0997400765 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 7 |
Hb_001288_050 |
0.1001102343 |
- |
- |
- |
| 8 |
Hb_000847_120 |
0.1059134832 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 9 |
Hb_001597_090 |
0.1263559715 |
- |
- |
- |
| 10 |
Hb_168319_060 |
0.1347433478 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 11 |
Hb_004943_010 |
0.1375204734 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_008376_020 |
0.1380604735 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 13 |
Hb_000751_050 |
0.1395887397 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 14 |
Hb_025668_020 |
0.1401169192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_110737_010 |
0.1407207159 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 16 |
Hb_002379_060 |
0.1412173151 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 17 |
Hb_007676_060 |
0.1426131733 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 18 |
Hb_000087_120 |
0.1491901162 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 19 |
Hb_011576_010 |
0.1493405287 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_003095_110 |
0.1508791312 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |