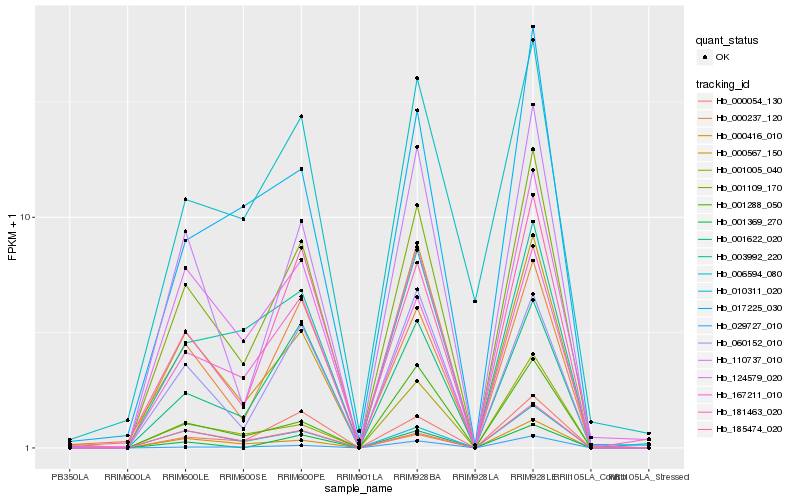
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001109_170 |
0.0 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_167211_010 |
0.0932233055 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_110737_010 |
0.0963064851 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 4 |
Hb_010311_020 |
0.10098575 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000416_010 |
0.1036233296 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 6 |
Hb_124579_020 |
0.1073547168 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 7 |
Hb_000054_130 |
0.1111236287 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 8 |
Hb_181463_020 |
0.1114054209 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 9 |
Hb_000567_150 |
0.1161326743 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 10 |
Hb_000237_120 |
0.1183189452 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 11 |
Hb_001005_040 |
0.1185675764 |
- |
- |
- |
| 12 |
Hb_001622_020 |
0.1199618959 |
- |
- |
- |
| 13 |
Hb_060152_010 |
0.1322582668 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 14 |
Hb_003992_220 |
0.1346189444 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 15 |
Hb_017225_030 |
0.1366633752 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 16 |
Hb_001288_050 |
0.1391558345 |
- |
- |
- |
| 17 |
Hb_006594_080 |
0.142237236 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 18 |
Hb_001369_270 |
0.1435554333 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_029727_010 |
0.1441883305 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_185474_020 |
0.1453161072 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |