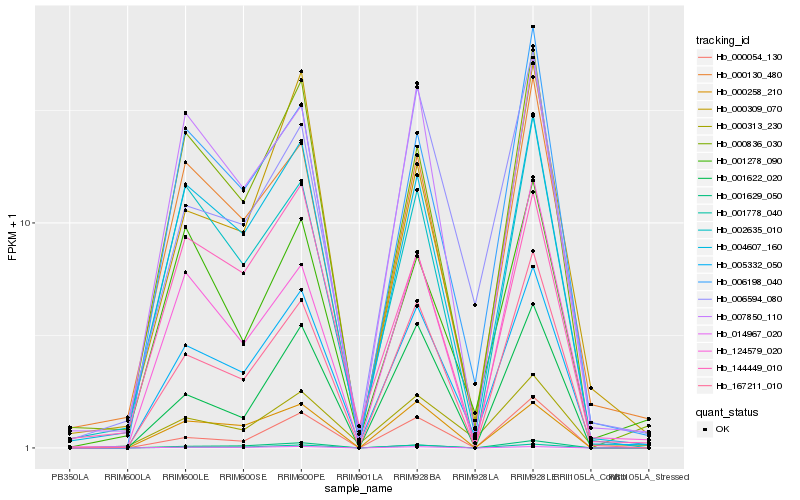
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 2 |
Hb_004607_160 |
0.0894301 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 3 |
Hb_167211_010 |
0.0991790214 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000130_480 |
0.113653712 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 5 |
Hb_007850_110 |
0.1155350674 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 6 |
Hb_000836_030 |
0.1169600398 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 7 |
Hb_001778_040 |
0.1179593226 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 8 |
Hb_000313_230 |
0.1193064064 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 9 |
Hb_006198_040 |
0.1206326662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001629_050 |
0.127475572 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 11 |
Hb_014967_020 |
0.1293910286 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_006594_080 |
0.1313818972 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_002635_010 |
0.1325068948 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 14 |
Hb_001622_020 |
0.1343147736 |
- |
- |
- |
| 15 |
Hb_000258_210 |
0.1343335625 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 16 |
Hb_000054_130 |
0.1343925112 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 17 |
Hb_001278_090 |
0.1362107785 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 18 |
Hb_124579_020 |
0.1384217502 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 19 |
Hb_000309_070 |
0.1391121965 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 20 |
Hb_144449_010 |
0.1393791277 |
- |
- |
Potassium transporter, putative [Ricinus communis] |