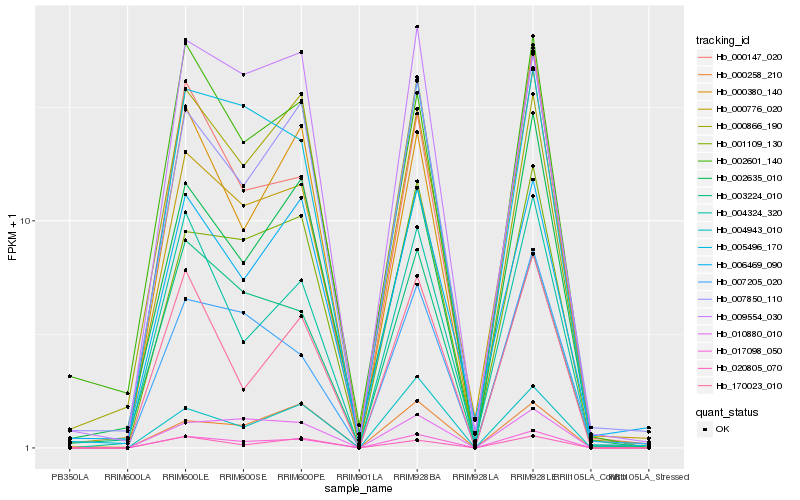
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017098_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 2 |
Hb_001109_130 |
0.0769279043 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007850_110 |
0.0844360573 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 4 |
Hb_005496_170 |
0.0951186045 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 5 |
Hb_010880_010 |
0.1040374741 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000776_020 |
0.1046767429 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 7 |
Hb_004943_010 |
0.1081238655 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000866_190 |
0.1183092806 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004324_320 |
0.1183729491 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_170023_010 |
0.1185918429 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 11 |
Hb_007205_020 |
0.1193235822 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 12 |
Hb_002601_140 |
0.1199206712 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 13 |
Hb_000147_020 |
0.1199384844 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 14 |
Hb_002635_010 |
0.1221784034 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 15 |
Hb_000258_210 |
0.1225483212 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 16 |
Hb_003224_010 |
0.1228225934 |
- |
- |
- |
| 17 |
Hb_009554_030 |
0.1236734899 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 18 |
Hb_006469_090 |
0.1258573401 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 19 |
Hb_020805_070 |
0.1260617627 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 20 |
Hb_000380_140 |
0.1261149005 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |