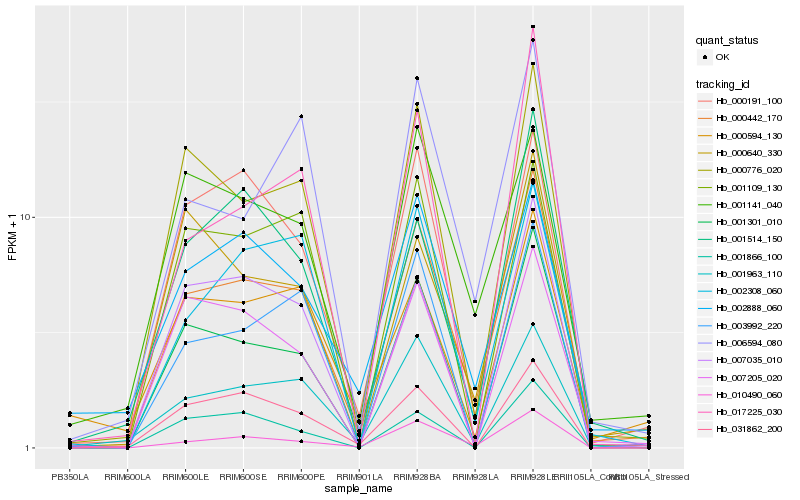
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_170 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_001301_010 |
0.1067287207 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000776_020 |
0.1068979415 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 4 |
Hb_001963_110 |
0.1194743033 |
- |
- |
- |
| 5 |
Hb_003992_220 |
0.1320809586 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 6 |
Hb_006594_080 |
0.1348705265 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 7 |
Hb_000191_100 |
0.1357264384 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_002308_060 |
0.1385606573 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 9 |
Hb_002888_060 |
0.1443828295 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031862_200 |
0.1466871629 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 11 |
Hb_001514_150 |
0.1473878898 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 12 |
Hb_017225_030 |
0.1483637087 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 13 |
Hb_007035_010 |
0.1506324752 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010490_060 |
0.1509368687 |
- |
- |
PREDICTED: uncharacterized protein LOC105767021 [Gossypium raimondii] |
| 15 |
Hb_001141_040 |
0.1511388664 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 16 |
Hb_000594_130 |
0.1545303887 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_001866_100 |
0.155196416 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 18 |
Hb_000640_330 |
0.1554306809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007205_020 |
0.1555156087 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 20 |
Hb_001109_130 |
0.1559571371 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |