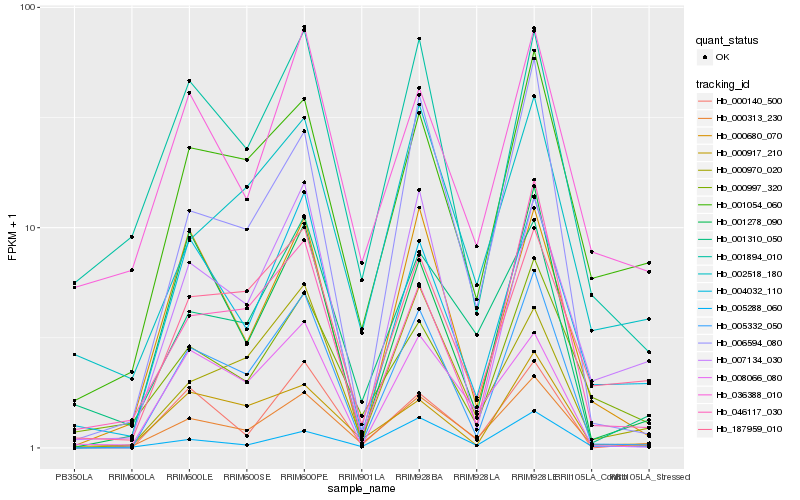
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 2 |
Hb_005332_050 |
0.1193064064 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 3 |
Hb_001054_060 |
0.1308557518 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 4 |
Hb_006594_080 |
0.1351628917 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 5 |
Hb_005288_060 |
0.1429598719 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_000997_320 |
0.1440242081 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 7 |
Hb_001310_050 |
0.1440797606 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 8 |
Hb_002518_180 |
0.1484716466 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 9 |
Hb_004032_110 |
0.1488171983 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 10 |
Hb_187959_010 |
0.1491344956 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 11 |
Hb_001894_010 |
0.1530828344 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 12 |
Hb_036388_010 |
0.1567151929 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 13 |
Hb_000680_070 |
0.1578919333 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001278_090 |
0.1597359054 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_000970_020 |
0.1614295763 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 16 |
Hb_007134_030 |
0.1629925107 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 17 |
Hb_008066_080 |
0.1630223161 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 18 |
Hb_000140_500 |
0.1638597383 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000917_210 |
0.1641825826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_046117_030 |
0.1651784014 |
- |
- |
sialyltransferase, putative [Ricinus communis] |