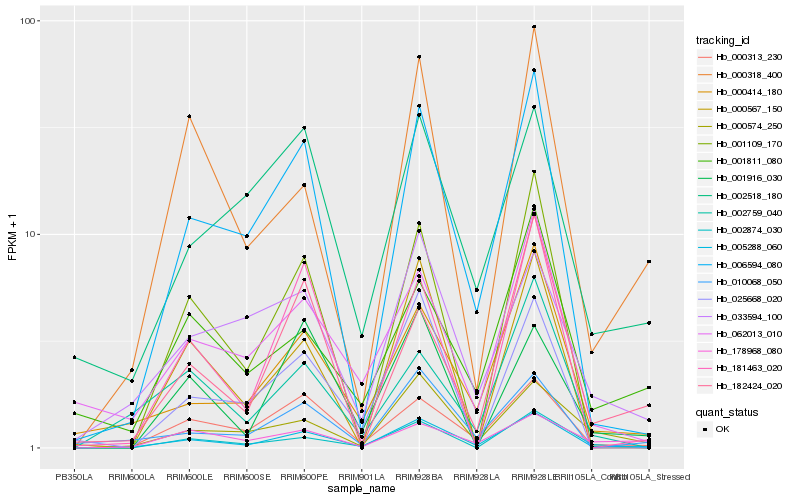
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_006594_080 |
0.1354538248 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 3 |
Hb_000313_230 |
0.1429598719 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 4 |
Hb_000414_180 |
0.1496228522 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 5 |
Hb_000574_250 |
0.1557412944 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 6 |
Hb_010068_050 |
0.1567891964 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 7 |
Hb_002874_030 |
0.1701931855 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 8 |
Hb_062013_010 |
0.1784294266 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000318_400 |
0.1821580924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_033594_100 |
0.1858138217 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001109_170 |
0.188692893 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002759_040 |
0.1944635761 |
- |
- |
BnaA09g29830D [Brassica napus] |
| 13 |
Hb_001811_080 |
0.1946165106 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 14 |
Hb_025668_020 |
0.1958135356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002518_180 |
0.1965222343 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 16 |
Hb_181463_020 |
0.196792203 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 17 |
Hb_182424_020 |
0.1971199572 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 18 |
Hb_001916_030 |
0.1981742114 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 19 |
Hb_000567_150 |
0.199183691 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_178968_080 |
0.199993034 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |