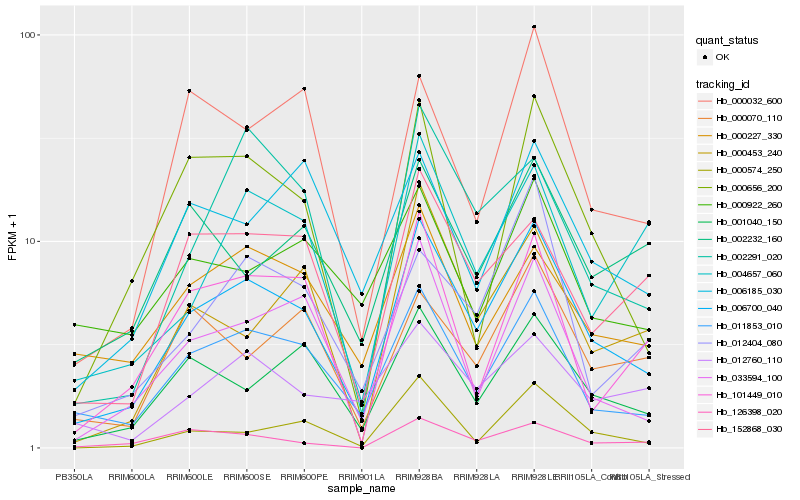
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006700_040 |
0.0 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 2 |
Hb_011853_010 |
0.1143388226 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 3 |
Hb_001040_150 |
0.1450208682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033594_100 |
0.1524328472 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_126398_020 |
0.1628897773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_152868_030 |
0.1644210523 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 7 |
Hb_000656_200 |
0.1646037493 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 8 |
Hb_000227_330 |
0.1699281549 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 9 |
Hb_002291_020 |
0.1708780419 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000032_600 |
0.1741701543 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_004657_060 |
0.1761217982 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_012760_110 |
0.1783733194 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000453_240 |
0.1788334957 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 14 |
Hb_006185_030 |
0.179698324 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 15 |
Hb_101449_010 |
0.1798199706 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 16 |
Hb_012404_080 |
0.1816931373 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 17 |
Hb_000070_110 |
0.1821939847 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002232_160 |
0.1822918631 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 19 |
Hb_000574_250 |
0.1843011755 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 20 |
Hb_000922_260 |
0.1846008135 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |