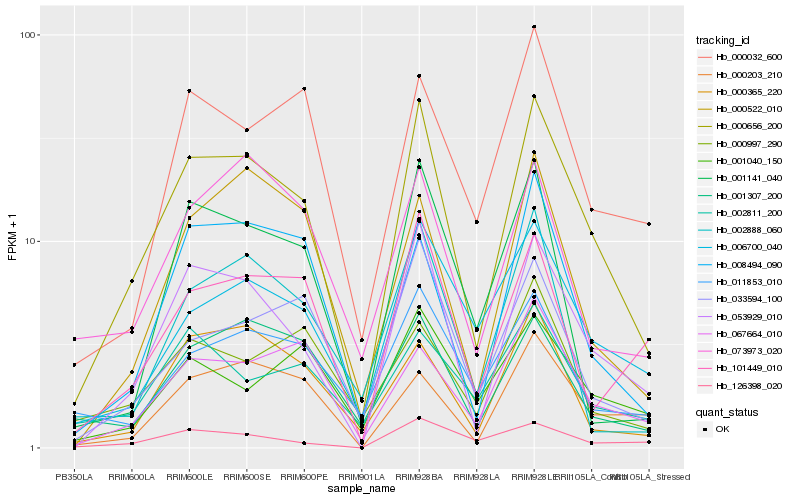
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 2 |
Hb_053929_010 |
0.1032124283 |
- |
- |
hypothetical protein POPTR_0010s22320g [Populus trichocarpa] |
| 3 |
Hb_033594_100 |
0.1627846521 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_006700_040 |
0.1646037493 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 5 |
Hb_008494_090 |
0.1675955866 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 6 |
Hb_067664_010 |
0.1681401776 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 7 |
Hb_000365_220 |
0.1746277074 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 8 |
Hb_000522_010 |
0.1765529792 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 9 |
Hb_101449_010 |
0.1768115888 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 10 |
Hb_001307_200 |
0.1779704727 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 11 |
Hb_000203_210 |
0.1789230249 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 12 |
Hb_011853_010 |
0.1797229904 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 13 |
Hb_000032_600 |
0.1805657299 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 14 |
Hb_002888_060 |
0.1809767147 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_126398_020 |
0.1814793945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001141_040 |
0.1814990795 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 17 |
Hb_073973_020 |
0.185943641 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 18 |
Hb_001040_150 |
0.1913410766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000997_290 |
0.1937256384 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 20 |
Hb_002811_200 |
0.195873593 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |