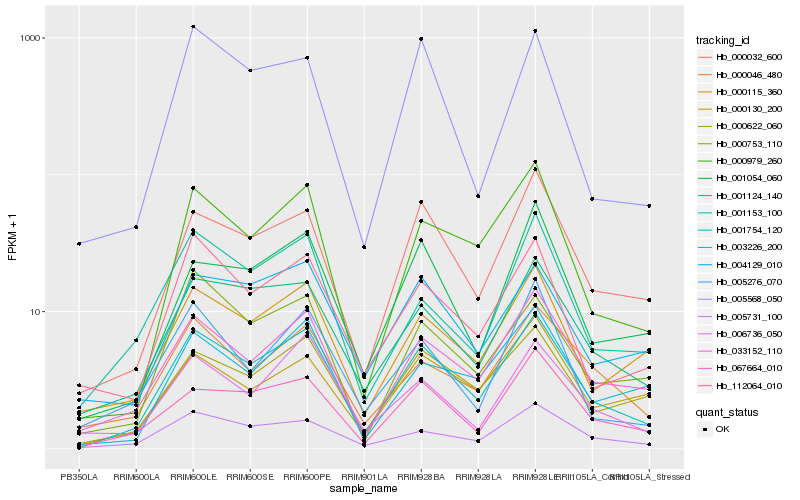
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_120 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 2 |
Hb_033152_110 |
0.1030046197 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 3 |
Hb_000032_600 |
0.1060843884 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_000979_260 |
0.1086758591 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 5 |
Hb_005731_100 |
0.1092078775 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 6 |
Hb_000622_060 |
0.1149577862 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001124_140 |
0.1149887013 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 8 |
Hb_067664_010 |
0.1169163426 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 9 |
Hb_000115_360 |
0.1304849507 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001054_060 |
0.1333350582 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 11 |
Hb_000046_480 |
0.1336048201 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 12 |
Hb_112064_010 |
0.1337033662 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 13 |
Hb_003226_200 |
0.1387287324 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 14 |
Hb_004129_010 |
0.1394340332 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_006736_050 |
0.1406663125 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 16 |
Hb_001153_100 |
0.1413451695 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 17 |
Hb_000753_110 |
0.1431550888 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 18 |
Hb_005568_050 |
0.1469970029 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 19 |
Hb_005276_070 |
0.1486887035 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 20 |
Hb_000130_200 |
0.1488704913 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |