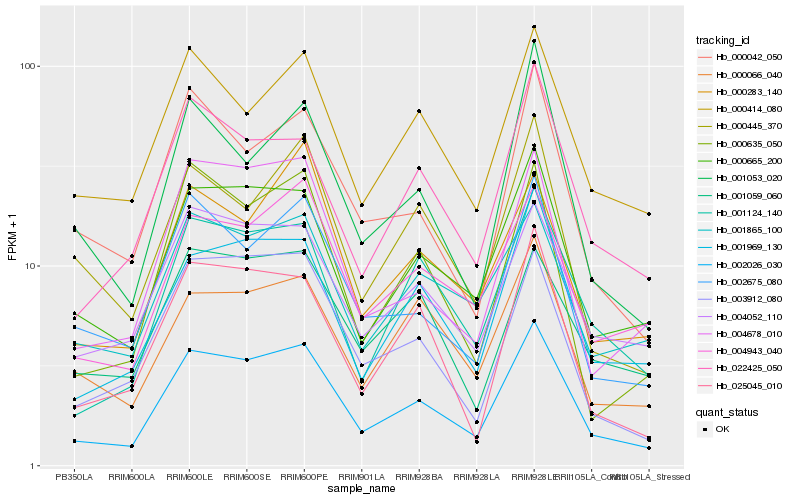
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_030 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 2 |
Hb_000665_200 |
0.0827565848 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_001969_130 |
0.0943275038 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 4 |
Hb_001124_140 |
0.0959817802 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 5 |
Hb_004052_110 |
0.1038391757 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002675_080 |
0.1047945039 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 7 |
Hb_004678_010 |
0.1050280127 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 8 |
Hb_003912_080 |
0.1069446651 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_004943_040 |
0.108843024 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 10 |
Hb_025045_010 |
0.1101149407 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_022425_050 |
0.1112339164 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 12 |
Hb_000635_050 |
0.1113737483 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 13 |
Hb_000414_080 |
0.1124149187 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000066_040 |
0.1133458392 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 15 |
Hb_000042_050 |
0.1160692646 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001865_100 |
0.1170663517 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 17 |
Hb_000445_370 |
0.1225013601 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 18 |
Hb_001059_060 |
0.1238852458 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 19 |
Hb_001053_020 |
0.124269874 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 20 |
Hb_000283_140 |
0.1257825301 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |