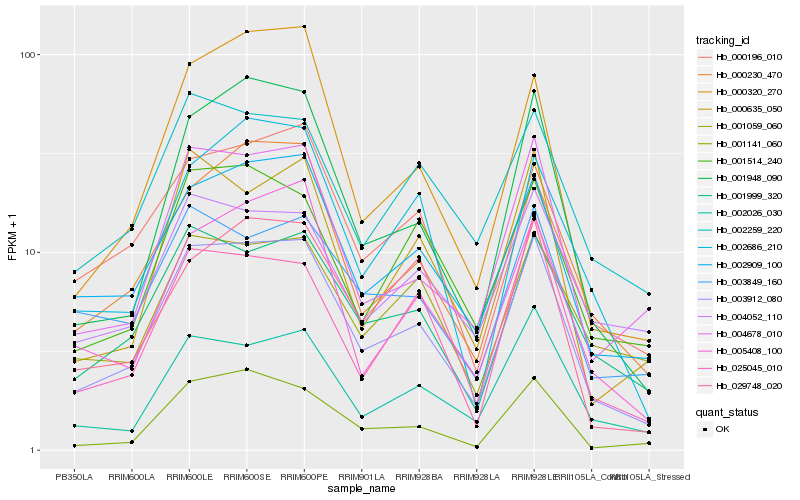
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003912_080 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 2 |
Hb_002909_100 |
0.0886062837 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 3 |
Hb_000230_470 |
0.0967476336 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001999_320 |
0.0967628433 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 5 |
Hb_004678_010 |
0.0994179147 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 6 |
Hb_001948_090 |
0.10059029 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 7 |
Hb_002026_030 |
0.1069446651 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 8 |
Hb_001141_060 |
0.1092521655 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_025045_010 |
0.1168205224 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000635_050 |
0.1174751733 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 11 |
Hb_005408_100 |
0.1192644973 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 12 |
Hb_003849_160 |
0.1222764197 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 13 |
Hb_001514_240 |
0.1228756357 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 14 |
Hb_000320_270 |
0.1229548105 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002259_220 |
0.1233710736 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 16 |
Hb_000196_010 |
0.1255278143 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 17 |
Hb_002686_210 |
0.1287156391 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_029748_020 |
0.1287508532 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_004052_110 |
0.1288024447 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001059_060 |
0.1298740057 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |