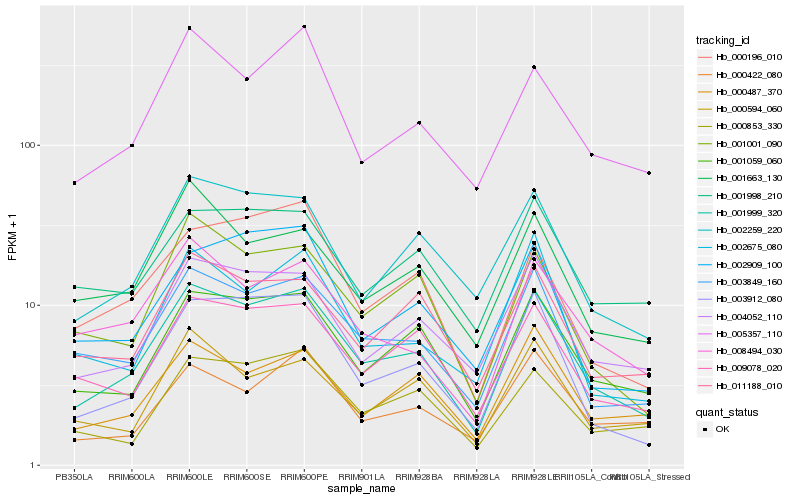
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_320 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 2 |
Hb_001059_060 |
0.0914522498 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 3 |
Hb_003849_160 |
0.0922602276 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 4 |
Hb_003912_080 |
0.0967628433 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 5 |
Hb_004052_110 |
0.1003904384 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002259_220 |
0.1075310321 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 7 |
Hb_009078_020 |
0.1075326646 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 8 |
Hb_001001_090 |
0.1137634371 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 9 |
Hb_000853_330 |
0.115873728 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_011188_010 |
0.1165009775 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005357_110 |
0.1165173461 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 12 |
Hb_008494_030 |
0.1218666022 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_000196_010 |
0.1231793606 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 14 |
Hb_000487_370 |
0.1232695859 |
- |
- |
- |
| 15 |
Hb_002909_100 |
0.1243802966 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 16 |
Hb_001998_210 |
0.1262165756 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000422_080 |
0.1278151542 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 18 |
Hb_001663_130 |
0.128387314 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 19 |
Hb_002675_080 |
0.1286917401 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 20 |
Hb_000594_060 |
0.1288811509 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |