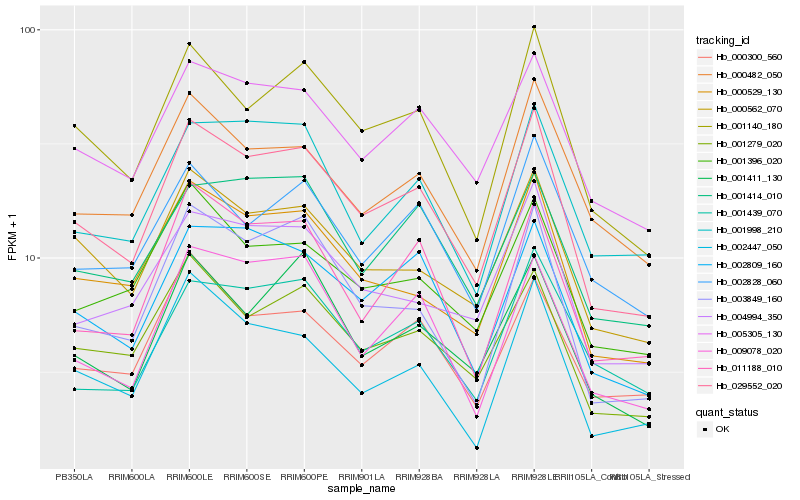
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029552_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001140_180 |
0.0614860627 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 3 |
Hb_001279_020 |
0.0716385917 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 4 |
Hb_005305_130 |
0.0726048806 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 5 |
Hb_000529_130 |
0.0732962061 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 6 |
Hb_002809_160 |
0.0751362452 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 7 |
Hb_000562_070 |
0.0760845211 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003849_160 |
0.0847766955 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 9 |
Hb_004994_350 |
0.0858426171 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000300_560 |
0.0873822839 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001396_020 |
0.088617628 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 12 |
Hb_011188_010 |
0.0886311296 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009078_020 |
0.0890111806 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 14 |
Hb_000482_050 |
0.0892440994 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001411_130 |
0.0905379935 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002447_050 |
0.0913187975 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001414_010 |
0.0927925753 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 18 |
Hb_001998_210 |
0.0937096212 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_002828_060 |
0.093733289 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001439_070 |
0.099890742 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |