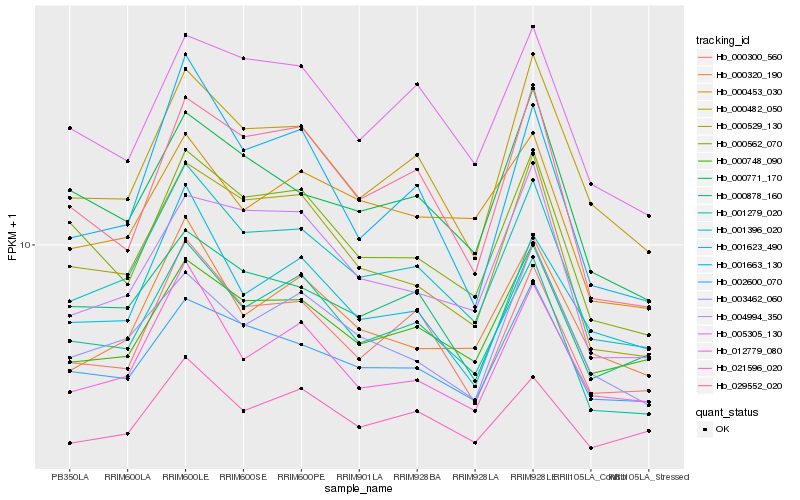
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_020 |
0.0 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 2 |
Hb_001279_020 |
0.0614650117 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 3 |
Hb_000300_560 |
0.0646205771 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000482_050 |
0.0720421768 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002600_070 |
0.0778663784 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 6 |
Hb_000529_130 |
0.0781457084 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 7 |
Hb_012779_080 |
0.0791710524 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000748_090 |
0.0796083408 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 9 |
Hb_000320_190 |
0.0810353261 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000562_070 |
0.0846467647 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021596_020 |
0.0857093141 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 12 |
Hb_000878_160 |
0.0864796956 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 13 |
Hb_005305_130 |
0.0868601828 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 14 |
Hb_029552_020 |
0.088617628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000771_170 |
0.0893861525 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_001623_490 |
0.090868389 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003462_060 |
0.091827004 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 18 |
Hb_004994_350 |
0.0923480509 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000453_030 |
0.0924853791 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 20 |
Hb_001663_130 |
0.0941456376 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |