| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
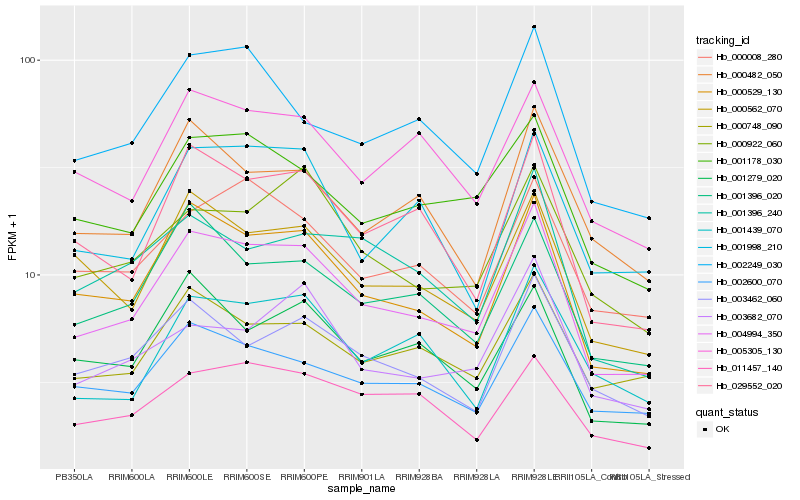
Hb_004994_350 |
0.0 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000529_130 |
0.0611650045 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 3 |
Hb_029552_020 |
0.0858426171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001178_030 |
0.0913713648 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 5 |
Hb_001396_020 |
0.0923480509 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_002600_070 |
0.0927266797 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 7 |
Hb_000748_090 |
0.0963046179 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 8 |
Hb_000562_070 |
0.096493342 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002249_030 |
0.0983305836 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003462_060 |
0.0983394517 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 11 |
Hb_000008_280 |
0.0988438062 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 12 |
Hb_005305_130 |
0.0989841404 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 13 |
Hb_000922_060 |
0.1006072333 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 14 |
Hb_001279_020 |
0.1023189665 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 15 |
Hb_000482_050 |
0.1035568701 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001439_070 |
0.1043445296 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001998_210 |
0.1045669415 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_003682_070 |
0.1062741935 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011457_140 |
0.1065534463 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 20 |
Hb_001396_240 |
0.1078969948 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |