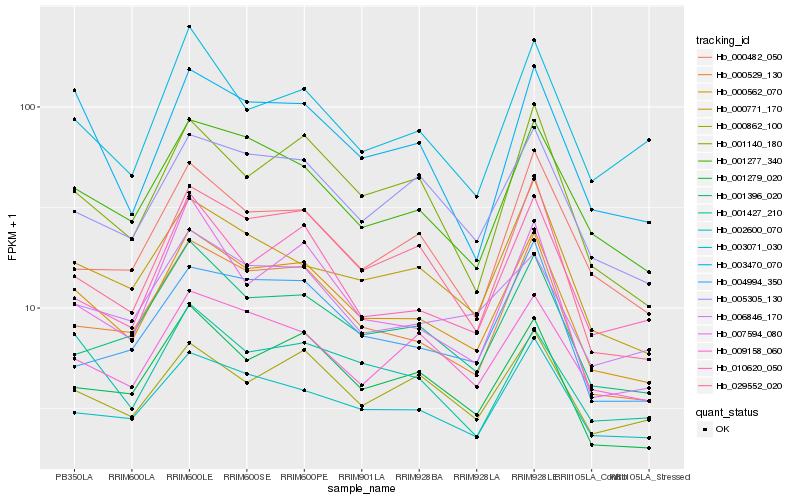
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000562_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000529_130 |
0.0618496674 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 3 |
Hb_001277_340 |
0.069818908 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 4 |
Hb_001279_020 |
0.0712797211 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 5 |
Hb_029552_020 |
0.0760845211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005305_130 |
0.0797755487 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 7 |
Hb_000771_170 |
0.0818143841 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_002600_070 |
0.0821820165 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 9 |
Hb_001396_020 |
0.0846467647 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 10 |
Hb_001140_180 |
0.0855179248 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 11 |
Hb_006846_170 |
0.0888671637 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 12 |
Hb_003470_070 |
0.0923777104 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 13 |
Hb_010620_050 |
0.093315604 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 14 |
Hb_009158_060 |
0.0953456909 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 15 |
Hb_000482_050 |
0.0956043316 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004994_350 |
0.096493342 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007594_080 |
0.0966098593 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 18 |
Hb_000862_100 |
0.0966636368 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001427_210 |
0.0978465868 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003071_030 |
0.1017640339 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |