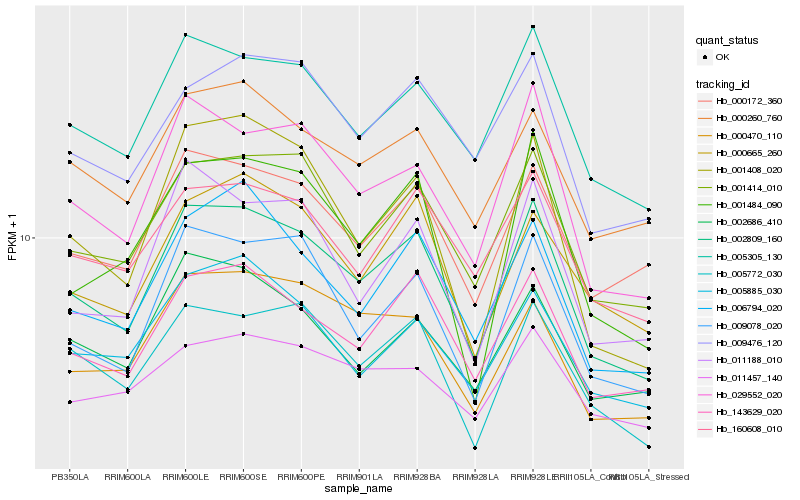
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002809_160 |
0.0 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 2 |
Hb_143629_020 |
0.0720938868 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 3 |
Hb_001414_010 |
0.073175077 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 4 |
Hb_029552_020 |
0.0751362452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005305_130 |
0.0782581172 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 6 |
Hb_009078_020 |
0.0802738777 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 7 |
Hb_000172_360 |
0.0877320063 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 8 |
Hb_006794_020 |
0.0882587303 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 9 |
Hb_001408_020 |
0.0916849893 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 10 |
Hb_005772_030 |
0.09205085 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 11 |
Hb_011188_010 |
0.093328974 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000260_760 |
0.0935386445 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 13 |
Hb_160608_010 |
0.094035463 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000665_260 |
0.0954196817 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011457_140 |
0.0964615235 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 16 |
Hb_009476_120 |
0.0966891713 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 17 |
Hb_005885_030 |
0.0968380999 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 18 |
Hb_001484_090 |
0.0971047454 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002686_410 |
0.097792399 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000470_110 |
0.0979759577 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |