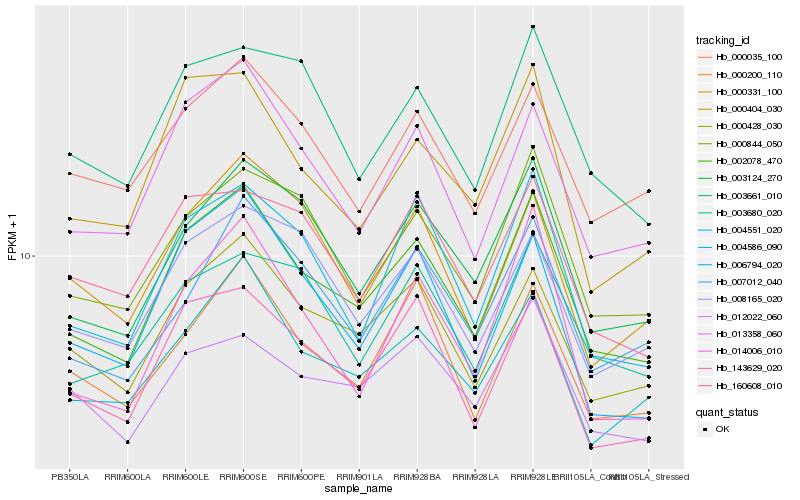
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_470 |
0.0 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000428_030 |
0.0638138883 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 3 |
Hb_013358_060 |
0.0680392074 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 4 |
Hb_003124_270 |
0.0699721004 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 5 |
Hb_004551_020 |
0.0715625517 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 6 |
Hb_143629_020 |
0.07368171 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 7 |
Hb_004586_090 |
0.075486565 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000844_050 |
0.0762523488 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 9 |
Hb_000404_030 |
0.0789782312 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 10 |
Hb_000200_110 |
0.0835144161 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003661_010 |
0.0874196266 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012022_060 |
0.0875083304 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007012_040 |
0.0882646655 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 14 |
Hb_003680_020 |
0.0893528972 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_006794_020 |
0.0896384263 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 16 |
Hb_000035_100 |
0.0896673595 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_014006_010 |
0.0896900476 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 18 |
Hb_008165_020 |
0.0896937037 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_160608_010 |
0.0899146969 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000331_100 |
0.0921809492 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |