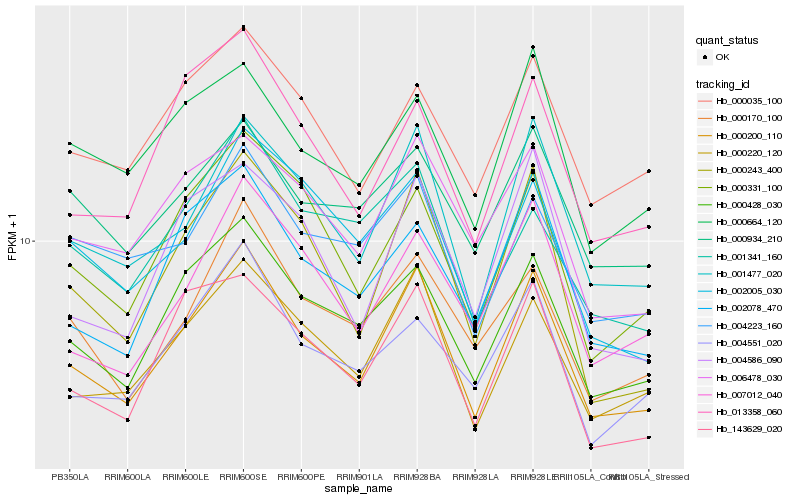
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_110 |
0.0 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001477_020 |
0.0591394239 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000220_120 |
0.0621377786 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000428_030 |
0.0682813667 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 5 |
Hb_000243_400 |
0.0764865866 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 6 |
Hb_000331_100 |
0.0793745997 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 7 |
Hb_143629_020 |
0.0807126636 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 8 |
Hb_000170_100 |
0.0832202015 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 9 |
Hb_002078_470 |
0.0835144161 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007012_040 |
0.0841722908 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 11 |
Hb_006478_030 |
0.0847774261 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 12 |
Hb_004586_090 |
0.0875479638 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004551_020 |
0.0886131502 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 14 |
Hb_000035_100 |
0.0893119828 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_013358_060 |
0.0893800842 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 16 |
Hb_000664_120 |
0.0930066499 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004223_160 |
0.0932137516 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002005_030 |
0.093584583 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_000934_210 |
0.095046705 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 20 |
Hb_001341_160 |
0.0951850821 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |