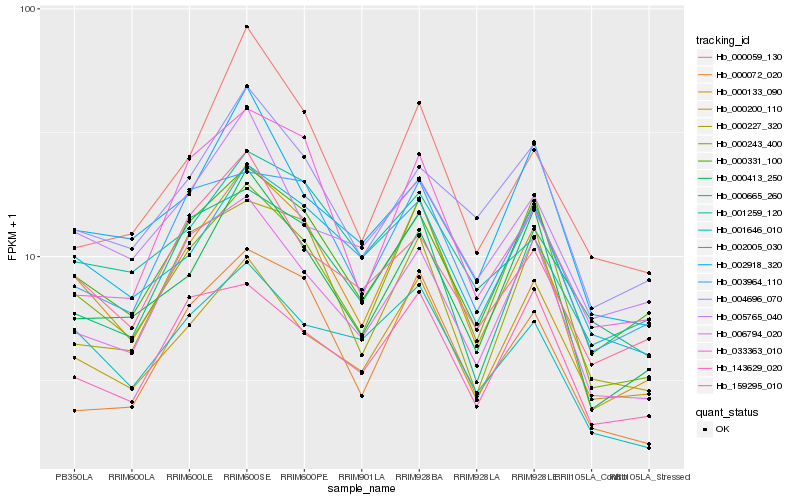
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000133_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 2 |
Hb_000243_400 |
0.0763741501 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 3 |
Hb_000413_250 |
0.0890782256 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 4 |
Hb_000331_100 |
0.0911043836 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006794_020 |
0.0947931196 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 6 |
Hb_000072_020 |
0.0989560494 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003964_110 |
0.0999281093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002005_030 |
0.1040079611 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_000227_320 |
0.1047488248 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001646_010 |
0.105488587 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 11 |
Hb_004696_070 |
0.1096685097 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 12 |
Hb_000200_110 |
0.111971593 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001259_120 |
0.1159872874 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 14 |
Hb_143629_020 |
0.1164555917 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 15 |
Hb_159295_010 |
0.1166013832 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 16 |
Hb_033363_010 |
0.116614767 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 17 |
Hb_002918_320 |
0.118153737 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_130 |
0.1199369024 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 19 |
Hb_000665_260 |
0.1199631842 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005765_040 |
0.1208190717 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |