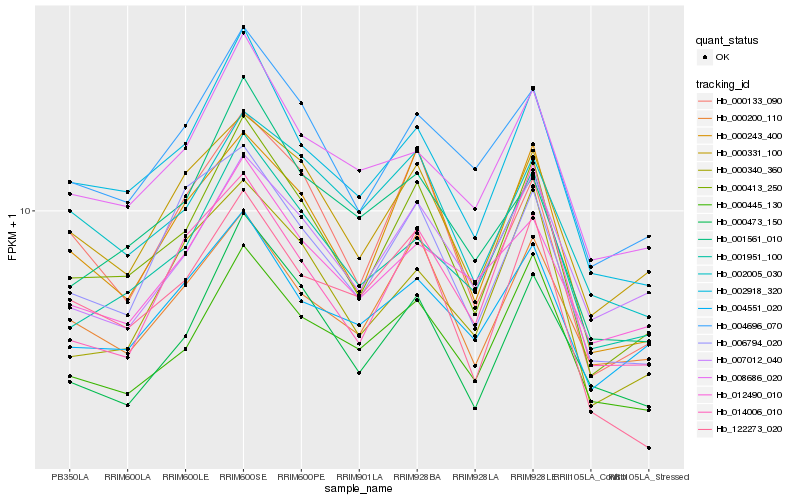
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_250 |
0.0 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 2 |
Hb_002918_320 |
0.0701030114 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000243_400 |
0.0780152208 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 4 |
Hb_004696_070 |
0.0856285418 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 5 |
Hb_001951_100 |
0.0867114374 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 6 |
Hb_000133_090 |
0.0890782256 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 7 |
Hb_000331_100 |
0.092449932 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000445_130 |
0.0933912904 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 9 |
Hb_000200_110 |
0.0978684188 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006794_020 |
0.0980156508 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 11 |
Hb_000473_150 |
0.0983773505 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008686_020 |
0.1016040873 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000340_360 |
0.1045929761 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 14 |
Hb_001561_010 |
0.1056062767 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004551_020 |
0.1057911266 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 16 |
Hb_007012_040 |
0.1073326941 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 17 |
Hb_002005_030 |
0.1109243855 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 18 |
Hb_012490_010 |
0.1114111687 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 19 |
Hb_122273_020 |
0.1124753882 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 20 |
Hb_014006_010 |
0.1138934804 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |