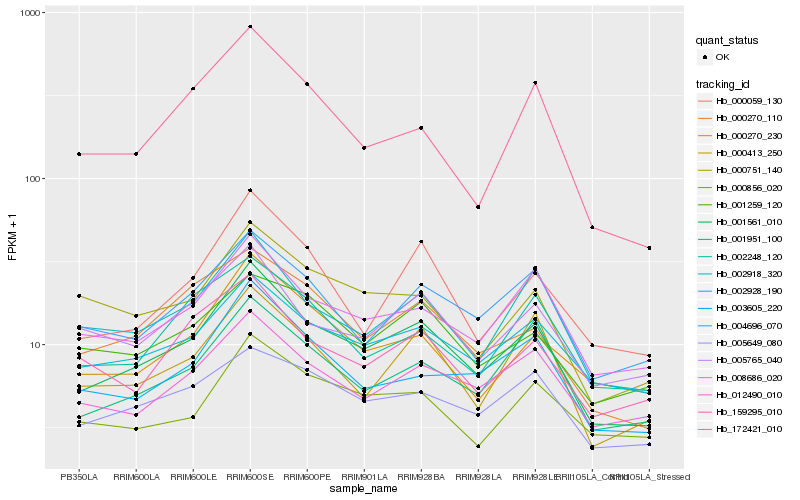
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001561_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002928_190 |
0.0908033295 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 3 |
Hb_002918_320 |
0.0914244349 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008686_020 |
0.0923335979 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001951_100 |
0.0983896362 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 6 |
Hb_004696_070 |
0.0993156091 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 7 |
Hb_003605_220 |
0.0993835331 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 8 |
Hb_000270_230 |
0.1000453963 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 9 |
Hb_012490_010 |
0.1020027484 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000270_110 |
0.1050114379 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 11 |
Hb_005765_040 |
0.1052818108 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 12 |
Hb_000413_250 |
0.1056062767 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 13 |
Hb_000059_130 |
0.1063254287 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 14 |
Hb_005649_080 |
0.108701522 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 15 |
Hb_000751_140 |
0.1121266963 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 16 |
Hb_000856_020 |
0.1122065062 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 17 |
Hb_159295_010 |
0.1137537758 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 18 |
Hb_002248_120 |
0.1137937349 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 19 |
Hb_001259_120 |
0.1143921986 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 20 |
Hb_172421_010 |
0.1167927238 |
- |
- |
cysteine protease [Hevea brasiliensis] |