| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
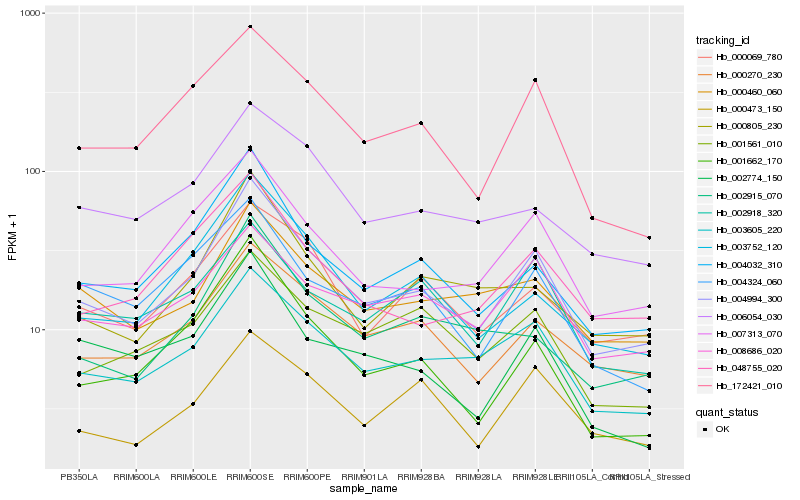
Hb_004994_300 |
0.0 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 2 |
Hb_003605_220 |
0.0834112863 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 3 |
Hb_007313_070 |
0.0936759465 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 4 |
Hb_000270_230 |
0.0966064115 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 5 |
Hb_172421_010 |
0.0974384526 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 6 |
Hb_004032_310 |
0.0997758071 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_003752_120 |
0.1104876844 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000460_060 |
0.1111388298 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 9 |
Hb_000069_780 |
0.1130345594 |
- |
- |
atpob1, putative [Ricinus communis] |
| 10 |
Hb_008686_020 |
0.115398234 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 11 |
Hb_048755_020 |
0.1158479714 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 12 |
Hb_006054_030 |
0.1163330922 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 13 |
Hb_000805_230 |
0.1183425267 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 14 |
Hb_002915_070 |
0.1195011975 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 15 |
Hb_004324_060 |
0.1201315795 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 16 |
Hb_001561_010 |
0.1201748552 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001662_170 |
0.1202572262 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 18 |
Hb_002918_320 |
0.1203099526 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000473_150 |
0.1207605239 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002774_150 |
0.1228905963 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |