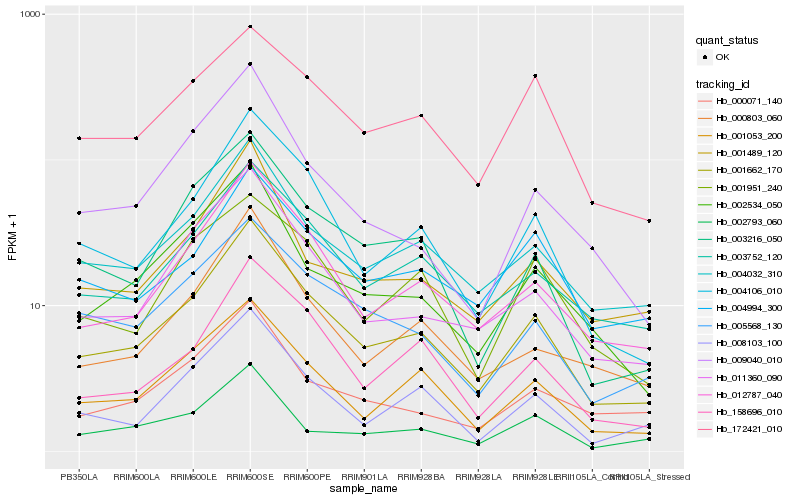
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_170 |
0.0 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 2 |
Hb_004106_010 |
0.0803762338 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012787_040 |
0.0806996974 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004032_310 |
0.0898620128 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 5 |
Hb_001053_200 |
0.0931594364 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 6 |
Hb_003752_120 |
0.0939032853 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011360_090 |
0.0984451896 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 8 |
Hb_158696_010 |
0.1063348574 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 9 |
Hb_008103_100 |
0.1070206852 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 10 |
Hb_002534_050 |
0.1087210943 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 11 |
Hb_003216_050 |
0.1107132836 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 12 |
Hb_005568_130 |
0.1128609997 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_001489_120 |
0.1132591798 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000803_060 |
0.1136698807 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 15 |
Hb_004994_300 |
0.1202572262 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 16 |
Hb_000071_140 |
0.1232070695 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 17 |
Hb_172421_010 |
0.1237591171 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 18 |
Hb_001951_240 |
0.1266210689 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 19 |
Hb_009040_010 |
0.1268047416 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_002793_060 |
0.1300082024 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |