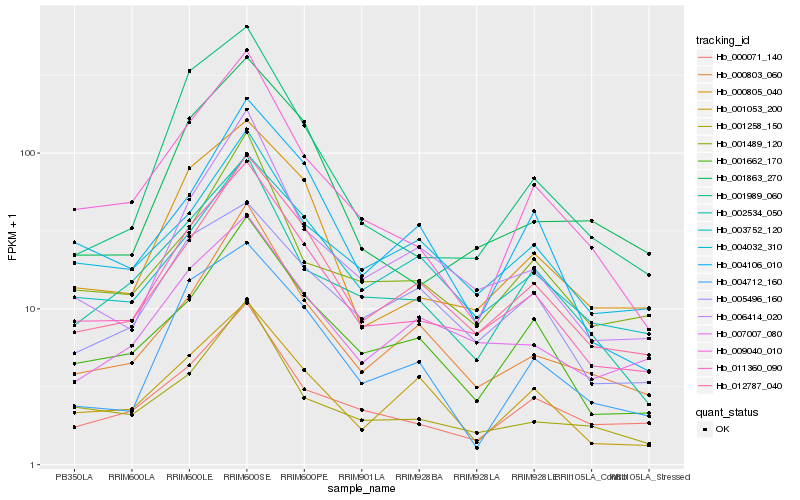
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011360_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 2 |
Hb_012787_040 |
0.0818796471 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000805_040 |
0.0916067096 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_001662_170 |
0.0984451896 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_009040_010 |
0.1030159876 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000803_060 |
0.1093442174 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 7 |
Hb_006414_020 |
0.1131007647 |
transcription factor |
TF Family: MYB |
- |
| 8 |
Hb_002534_050 |
0.1134922237 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 9 |
Hb_000071_140 |
0.1139123547 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 10 |
Hb_001258_150 |
0.1160759189 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 11 |
Hb_003752_120 |
0.1185499108 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007007_080 |
0.1206499304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004032_310 |
0.1218572029 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 14 |
Hb_001863_270 |
0.1228529698 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 15 |
Hb_001489_120 |
0.1242132741 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004106_010 |
0.1286008836 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001053_200 |
0.1300811697 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 18 |
Hb_001989_060 |
0.1318893958 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004712_160 |
0.1319543438 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 20 |
Hb_005496_160 |
0.1351933308 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |