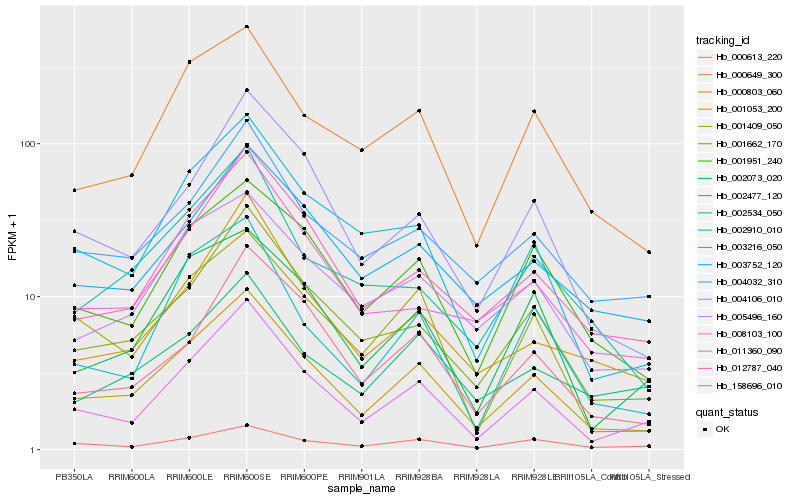
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_200 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 2 |
Hb_008103_100 |
0.0868059343 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 3 |
Hb_001662_170 |
0.0931594364 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 4 |
Hb_001951_240 |
0.0999133995 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 5 |
Hb_000649_300 |
0.1050700068 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 6 |
Hb_005496_160 |
0.1094767805 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004032_310 |
0.1096338717 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 8 |
Hb_003752_120 |
0.1106958329 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004106_010 |
0.1138925399 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012787_040 |
0.1145680671 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002073_020 |
0.1186183294 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 12 |
Hb_003216_050 |
0.1187258543 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 13 |
Hb_001409_050 |
0.1233715557 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_158696_010 |
0.125622381 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 15 |
Hb_000613_220 |
0.127734967 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 16 |
Hb_002910_010 |
0.1280662427 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 17 |
Hb_011360_090 |
0.1300811697 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 18 |
Hb_000803_060 |
0.1302815944 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 19 |
Hb_002477_120 |
0.132087052 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_002534_050 |
0.1335006715 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |