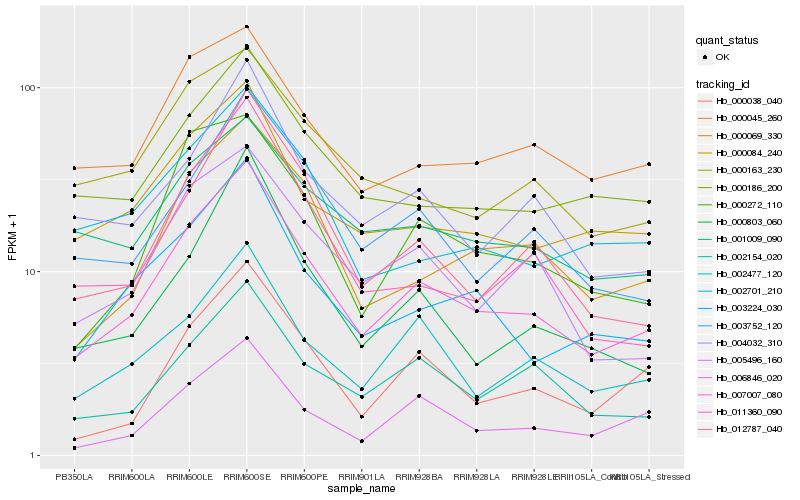
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007007_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000069_330 |
0.1023660302 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_003224_030 |
0.1043963458 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 4 |
Hb_003752_120 |
0.1082681796 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002477_120 |
0.1096980532 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_005496_160 |
0.110954935 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002154_020 |
0.113455003 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_012787_040 |
0.1136403912 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000272_110 |
0.1142096404 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004032_310 |
0.1145013012 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 11 |
Hb_000084_240 |
0.1154420059 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 12 |
Hb_000186_200 |
0.1162281148 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_011360_090 |
0.1206499304 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 14 |
Hb_000045_260 |
0.1217103086 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000803_060 |
0.123006521 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 16 |
Hb_000038_040 |
0.1240217275 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 17 |
Hb_006846_020 |
0.1244369601 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 18 |
Hb_000163_230 |
0.1281813595 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 19 |
Hb_002701_210 |
0.1293267041 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 20 |
Hb_001009_090 |
0.1299561021 |
- |
- |
CDK protein [Hevea brasiliensis] |