| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
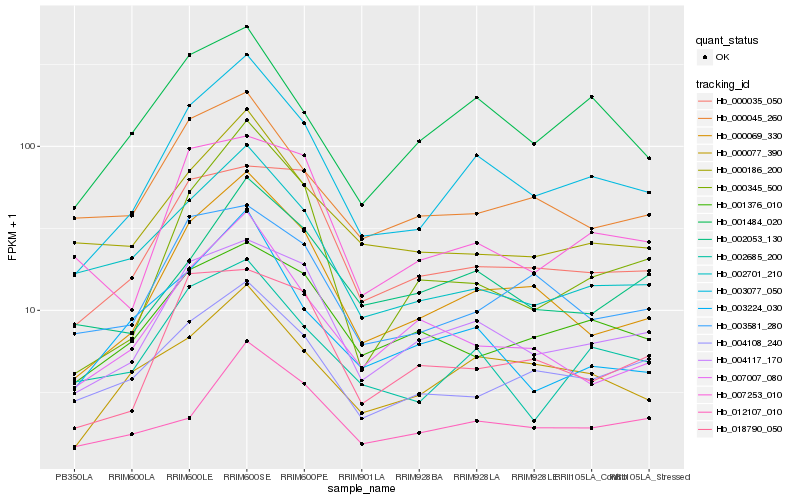
Hb_003077_050 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 2 |
Hb_000069_330 |
0.0886675842 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_004117_170 |
0.1103761424 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 4 |
Hb_000077_390 |
0.1207695415 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0019s13330g [Populus trichocarpa] |
| 5 |
Hb_002685_200 |
0.1239126529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 6 |
Hb_001484_020 |
0.1325491238 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000186_200 |
0.1364747839 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 8 |
Hb_004108_240 |
0.1373721674 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 9 |
Hb_002701_210 |
0.137689593 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 10 |
Hb_000045_260 |
0.1425362669 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007007_080 |
0.1428968674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003581_280 |
0.143958784 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 13 |
Hb_000345_500 |
0.1441298327 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 14 |
Hb_003224_030 |
0.144198754 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 15 |
Hb_012107_010 |
0.1474293632 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 16 |
Hb_002053_130 |
0.1474734139 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 17 |
Hb_001376_010 |
0.1481380223 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 18 |
Hb_000035_050 |
0.1486457978 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 19 |
Hb_007253_010 |
0.149844829 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 20 |
Hb_018790_050 |
0.1503200325 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |