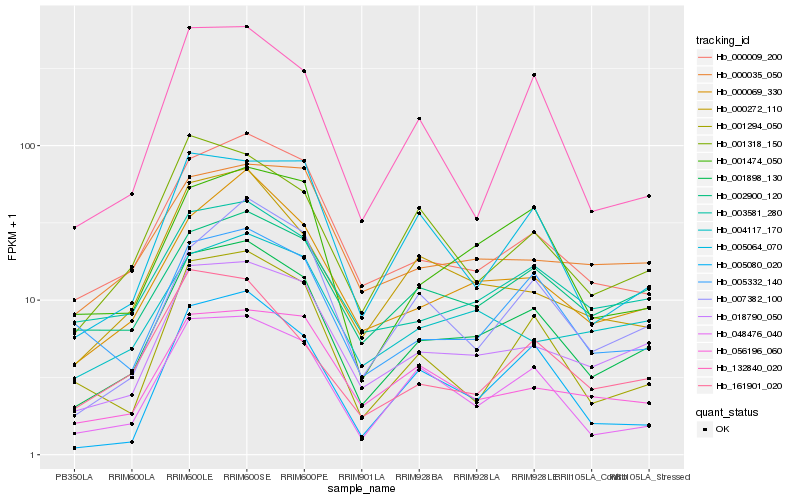
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_130 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_018790_050 |
0.0882422322 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 3 |
Hb_000069_330 |
0.1050702725 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_003581_280 |
0.1110738671 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 5 |
Hb_000009_200 |
0.113242227 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 6 |
Hb_001474_050 |
0.1140637094 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 7 |
Hb_048476_040 |
0.1174989396 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 8 |
Hb_002900_120 |
0.121423544 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 9 |
Hb_000272_110 |
0.125476298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007382_100 |
0.1265177262 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005332_140 |
0.1282751084 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001294_050 |
0.1305562297 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 13 |
Hb_005064_070 |
0.1317939521 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 14 |
Hb_004117_170 |
0.1319430812 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 15 |
Hb_001318_150 |
0.133913958 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 16 |
Hb_056196_060 |
0.1352606762 |
- |
- |
hexokinase [Manihot esculenta] |
| 17 |
Hb_161901_020 |
0.1373152456 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 18 |
Hb_000035_050 |
0.1378266005 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 19 |
Hb_132840_020 |
0.1380555677 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 20 |
Hb_005080_020 |
0.1406925691 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |