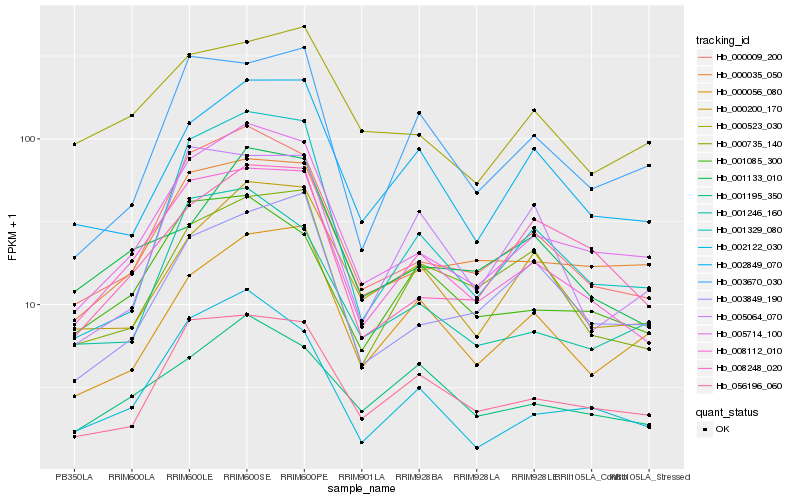
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008112_010 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 2 |
Hb_000009_200 |
0.0959793069 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 3 |
Hb_056196_060 |
0.1023344635 |
- |
- |
hexokinase [Manihot esculenta] |
| 4 |
Hb_005714_100 |
0.1032567827 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 5 |
Hb_000035_050 |
0.1118401013 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 6 |
Hb_001085_300 |
0.1134164475 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 7 |
Hb_000735_140 |
0.1203103145 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 8 |
Hb_003670_030 |
0.1276831613 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 9 |
Hb_002849_070 |
0.1304077285 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000523_030 |
0.1336198705 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 11 |
Hb_001329_080 |
0.1386181188 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 12 |
Hb_001195_350 |
0.1386702985 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 13 |
Hb_002122_030 |
0.1398477961 |
- |
- |
- |
| 14 |
Hb_005064_070 |
0.1408294756 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 15 |
Hb_000200_170 |
0.1415720294 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 16 |
Hb_001133_010 |
0.1437781676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000056_080 |
0.1447579296 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001246_160 |
0.1455710477 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 19 |
Hb_008248_020 |
0.1460792456 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_003849_190 |
0.1464555488 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |