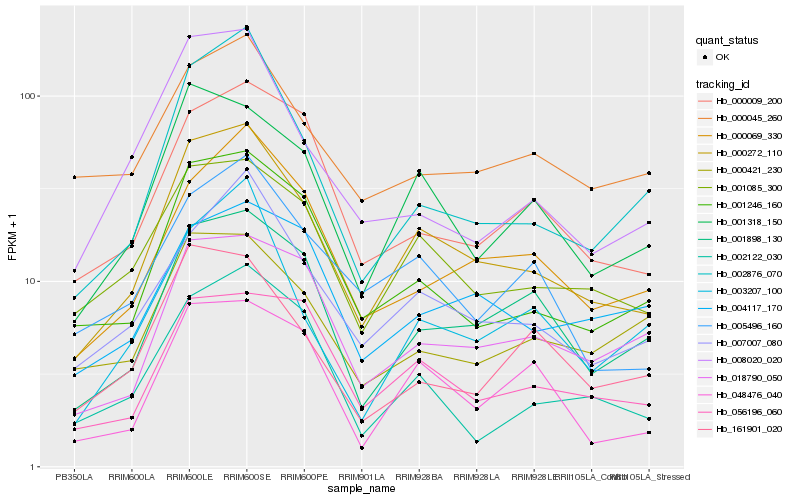
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000272_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001085_300 |
0.1102059283 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 3 |
Hb_007007_080 |
0.1142096404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001318_150 |
0.1143071582 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 5 |
Hb_001246_160 |
0.1175482232 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 6 |
Hb_001898_130 |
0.125476298 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_002876_070 |
0.1259989552 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 8 |
Hb_000069_330 |
0.1312057218 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_005496_160 |
0.1350625313 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000009_200 |
0.1385267084 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 11 |
Hb_018790_050 |
0.1395021444 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 12 |
Hb_056196_060 |
0.1416274618 |
- |
- |
hexokinase [Manihot esculenta] |
| 13 |
Hb_048476_040 |
0.1420515489 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 14 |
Hb_002122_030 |
0.1447909716 |
- |
- |
- |
| 15 |
Hb_003207_100 |
0.1482416512 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 16 |
Hb_000045_260 |
0.1487046588 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_008020_020 |
0.1511629423 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_161901_020 |
0.1520259763 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 19 |
Hb_004117_170 |
0.1546210974 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 20 |
Hb_000421_230 |
0.155607987 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |